2 ShiNyP
📄 Input data: Genome-wide biallelic SNP in Variant Call Format (VCF) file format.
📊 Analysis: Data QC, population genetics analysis, core collection…
📋 Output: Publication-ready figures, tables, analyzed data objects, and AI-driven report.
📊 Analysis: Data QC, population genetics analysis, core collection…
📋 Output: Publication-ready figures, tables, analyzed data objects, and AI-driven report.
Key Features
> Statistical and computational exploration
> Customizable visualization options
> Publication-ready figures and tables
> Analyzed R serialization objects
> Auto-generate preliminary results
> AI-driven report - powered by OpenAI
> Statistical and computational exploration
> Customizable visualization options
> Publication-ready figures and tables
> Analyzed R serialization objects
> Auto-generate preliminary results
> AI-driven report - powered by OpenAI
Publication
Huang et al. (upcoming 2024) ShiNyP: An Interactive Shiny-Based Platform for Genome-Wide SNP Analysis and Visualization
Support
If you encounter any issues or have suggestions for new features, please submit a report through our feedback form: https://forms.gle/GPCggSo5czyNLfoB7
Websites
Journal Article: Under Review…
GitHub Repository: https://github.com/TeddYenn/ShiNyP
User Manual: https://teddyenn.github.io/ShiNyP-guide
Demo Datasets: https://reurl.cc/QEx5lZ
Demo Platform: https://teddyhuang.shinyapps.io/ShiNyP
Feedback Form: https://forms.gle/GPCggSo5czyNLfoB7
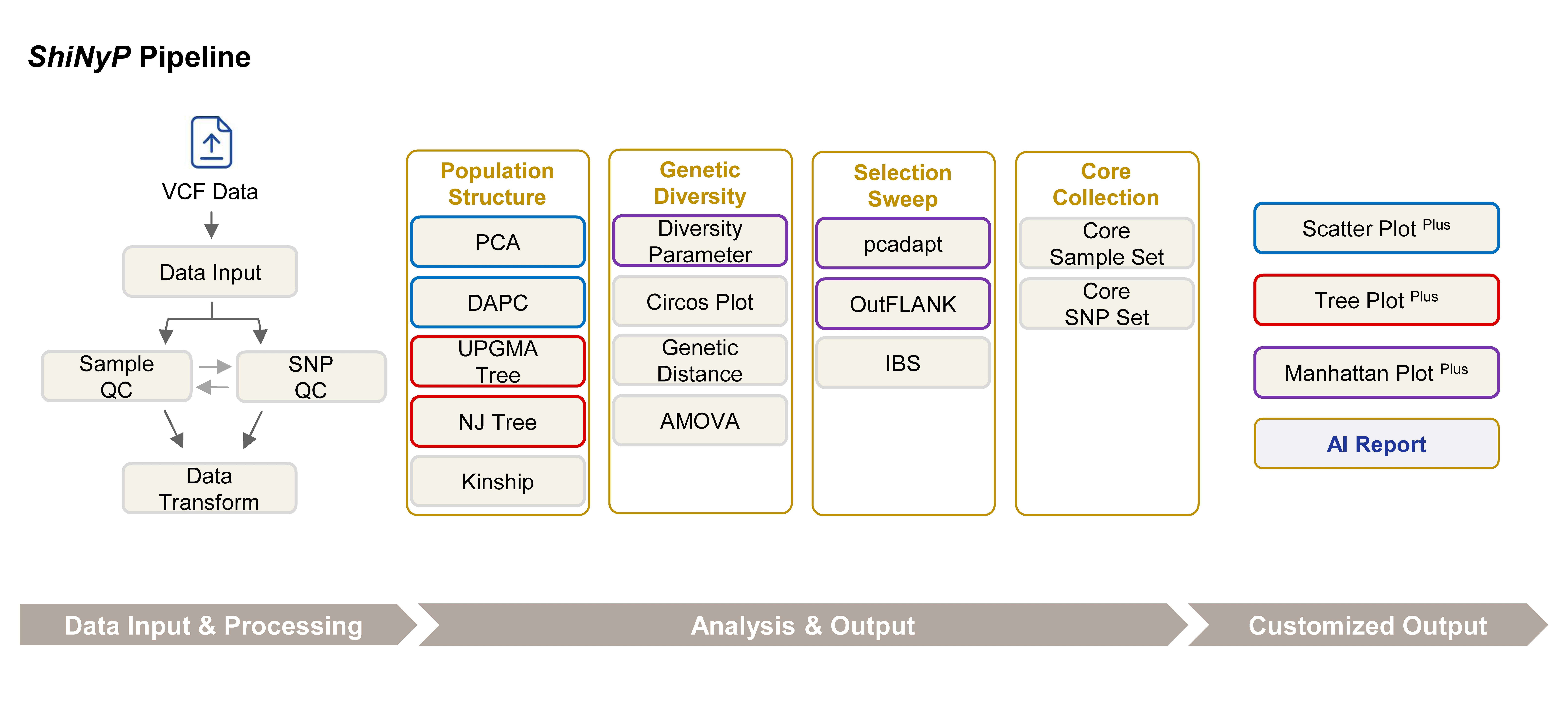
▲ An overview of the ShiNyP platform’s workflow for genome-wide SNP data analysis.
- Data Input & Processing: Beginning with VCF data input, it performs quality control (QC) and data transformation steps.
- Analysis & Output: The analysis phase is divided into modules, each represented as a page in the platform, with multiple subpages offering specific analytical functions, including population structure, genetic diversity, selection sweep, and core collection.
- Customized Output: The final output offers tailored visualizations and includes an AI-generated report summarizing the results. The pipeline streamlines data input, processing, and advanced analysis to deliver publication-ready figures and reports customized to the user’s needs.
*Subpage frame colors indicate available customizable functions. For example, blue frames for PCA and DAPC correspond to the Scatter Plot Plus tool for customizing scatter plots, while red and purple frames correspond to Tree Plot Plus and Manhattan Plot Plus, respectively.
- Data Input & Processing: Beginning with VCF data input, it performs quality control (QC) and data transformation steps.
- Analysis & Output: The analysis phase is divided into modules, each represented as a page in the platform, with multiple subpages offering specific analytical functions, including population structure, genetic diversity, selection sweep, and core collection.
- Customized Output: The final output offers tailored visualizations and includes an AI-generated report summarizing the results. The pipeline streamlines data input, processing, and advanced analysis to deliver publication-ready figures and reports customized to the user’s needs.
*Subpage frame colors indicate available customizable functions. For example, blue frames for PCA and DAPC correspond to the Scatter Plot Plus tool for customizing scatter plots, while red and purple frames correspond to Tree Plot Plus and Manhattan Plot Plus, respectively.