6 Selection Sweep
➡️ This section includes four subpages: pcadapt, OutFLANK, IBS, and Manhattan PlotPlus, allowing you to detect selection signatures in different scenario and customize your plot.
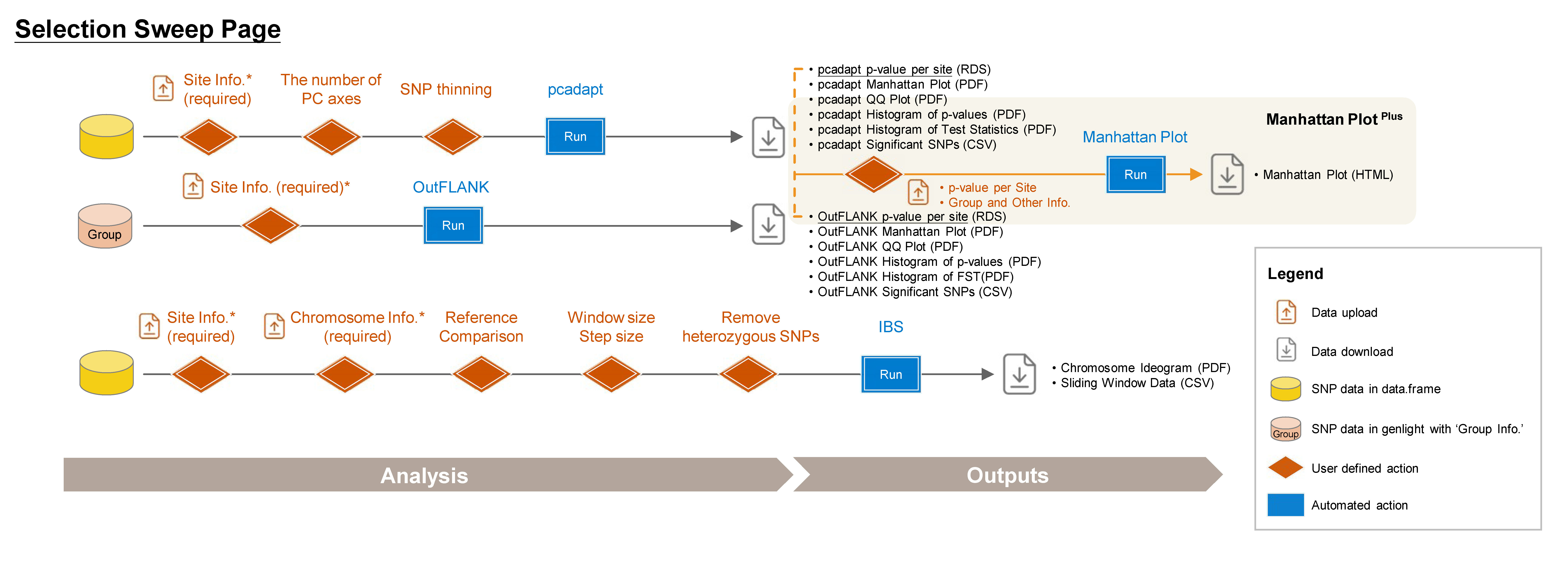
6.1 pcadapt
A PCA-based approach identifies selective outliers relative to population structure (Luu, Bazin, and Blum 2016).
Required Files:
- data.frame
- Site Info. (RDS) of the current data.frame, downloadable from Data Input or Data QC pages.
Steps:
- Upload Site Info. (required).
- Click SNP Thinning (optional) and choose window size (number of SNPs) and r² threshold. For more information, visit https://bcm-uga.github.io/pcadapt/articles/pcadapt.html.
- Click Run pcadapt to perform genome scan for selection.
Outputs:
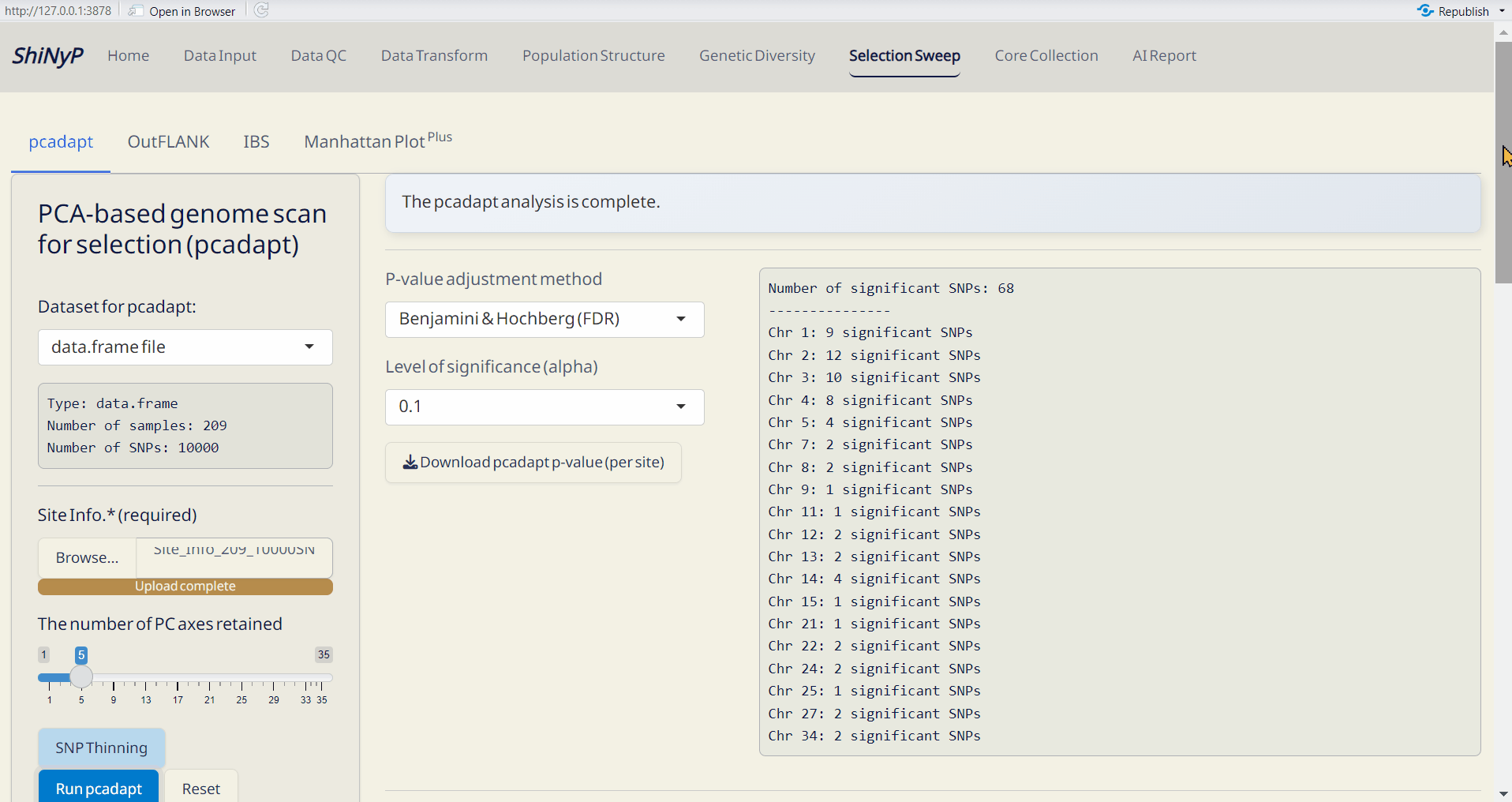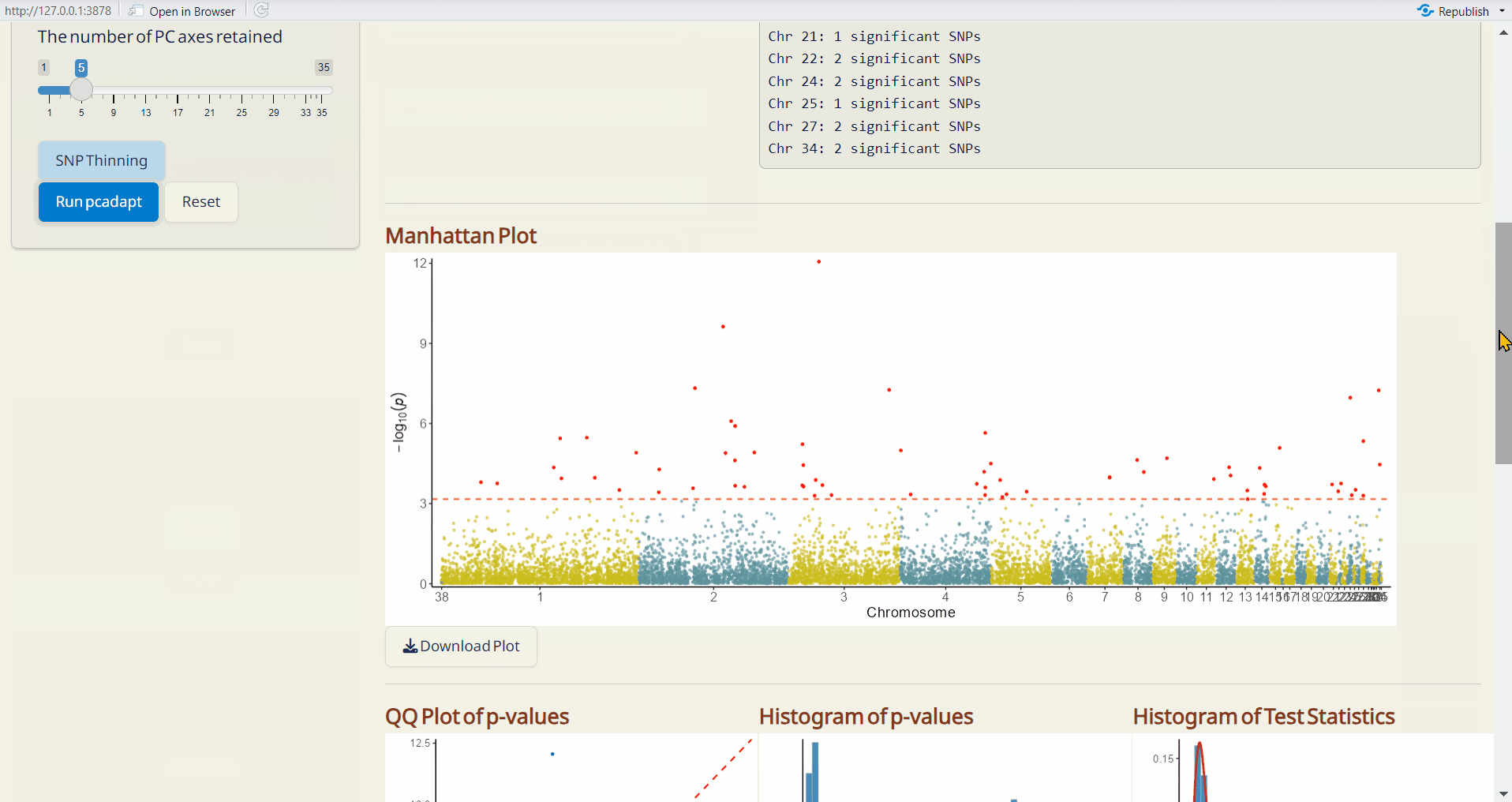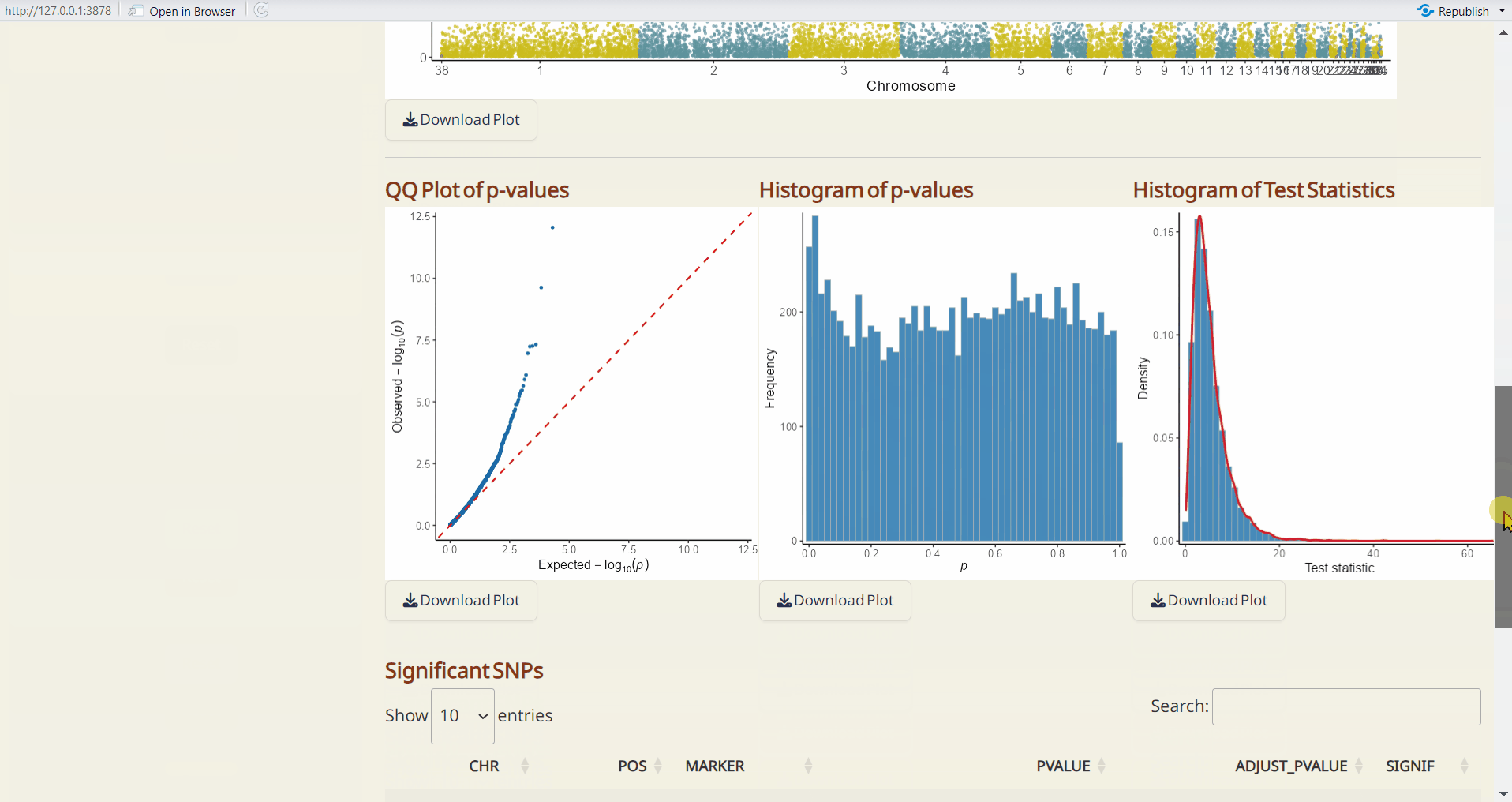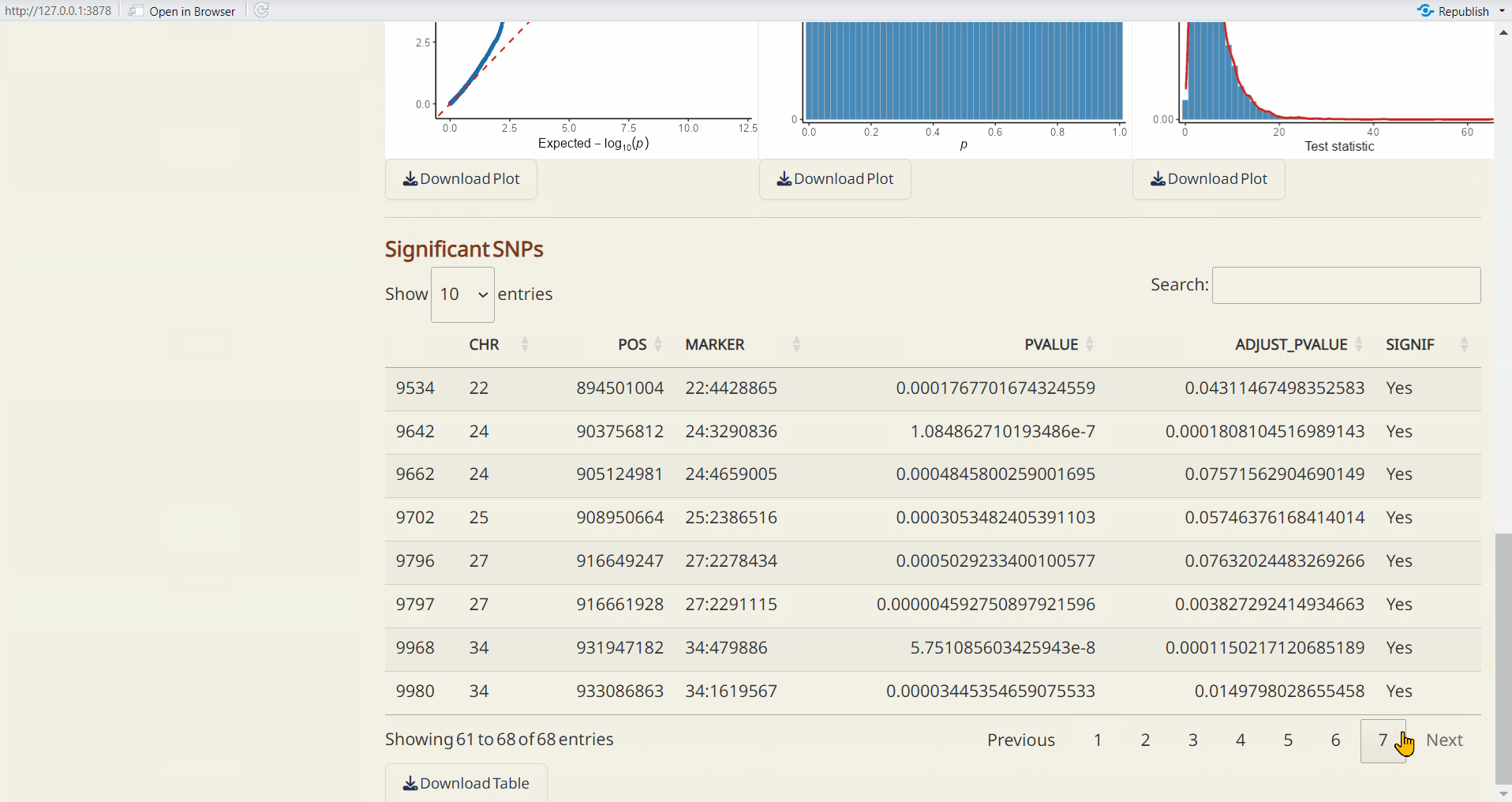
- pcadapt p-value per site (RDS): A dataset containing p-values and adjusted p-values for each site.
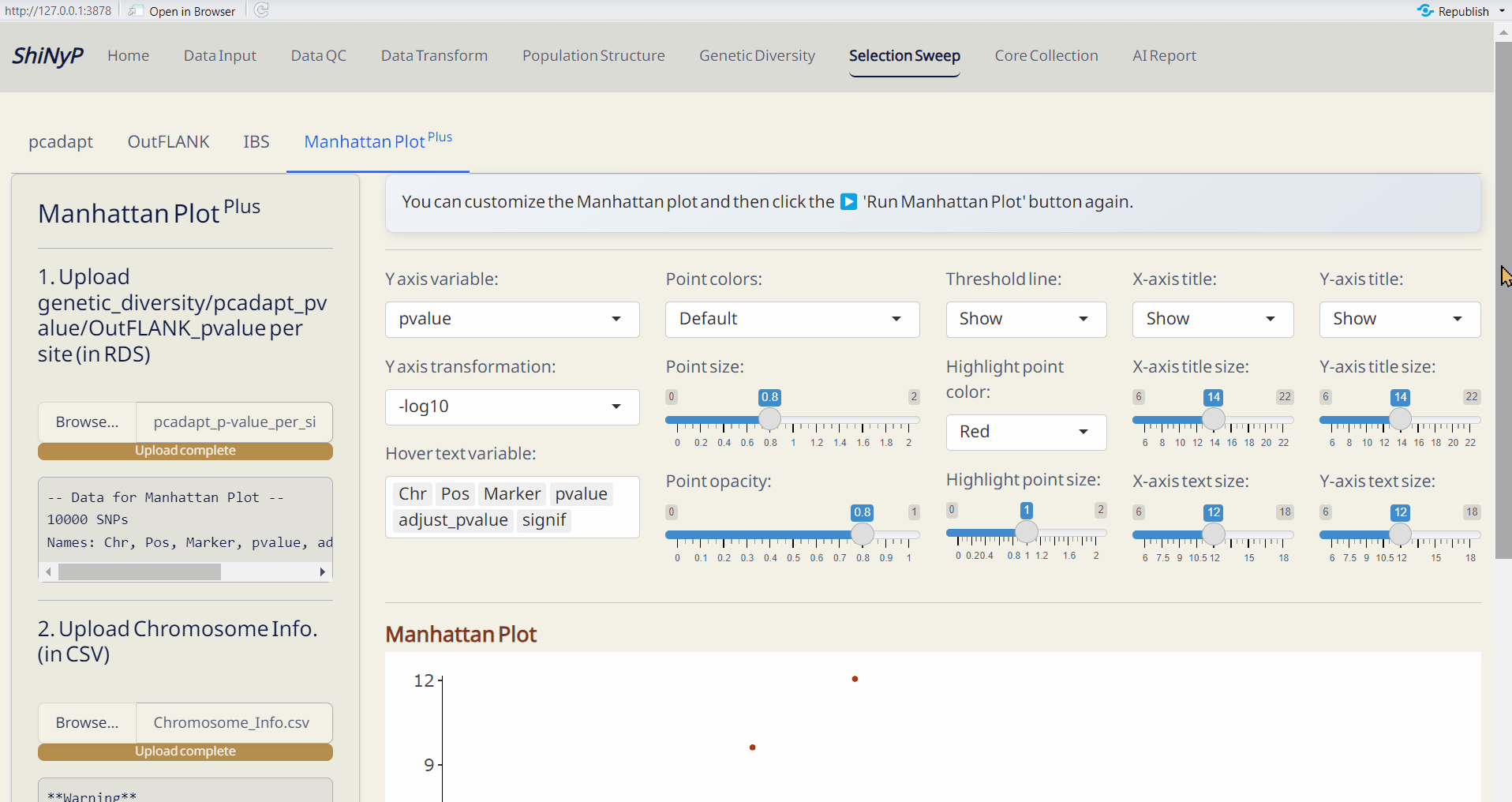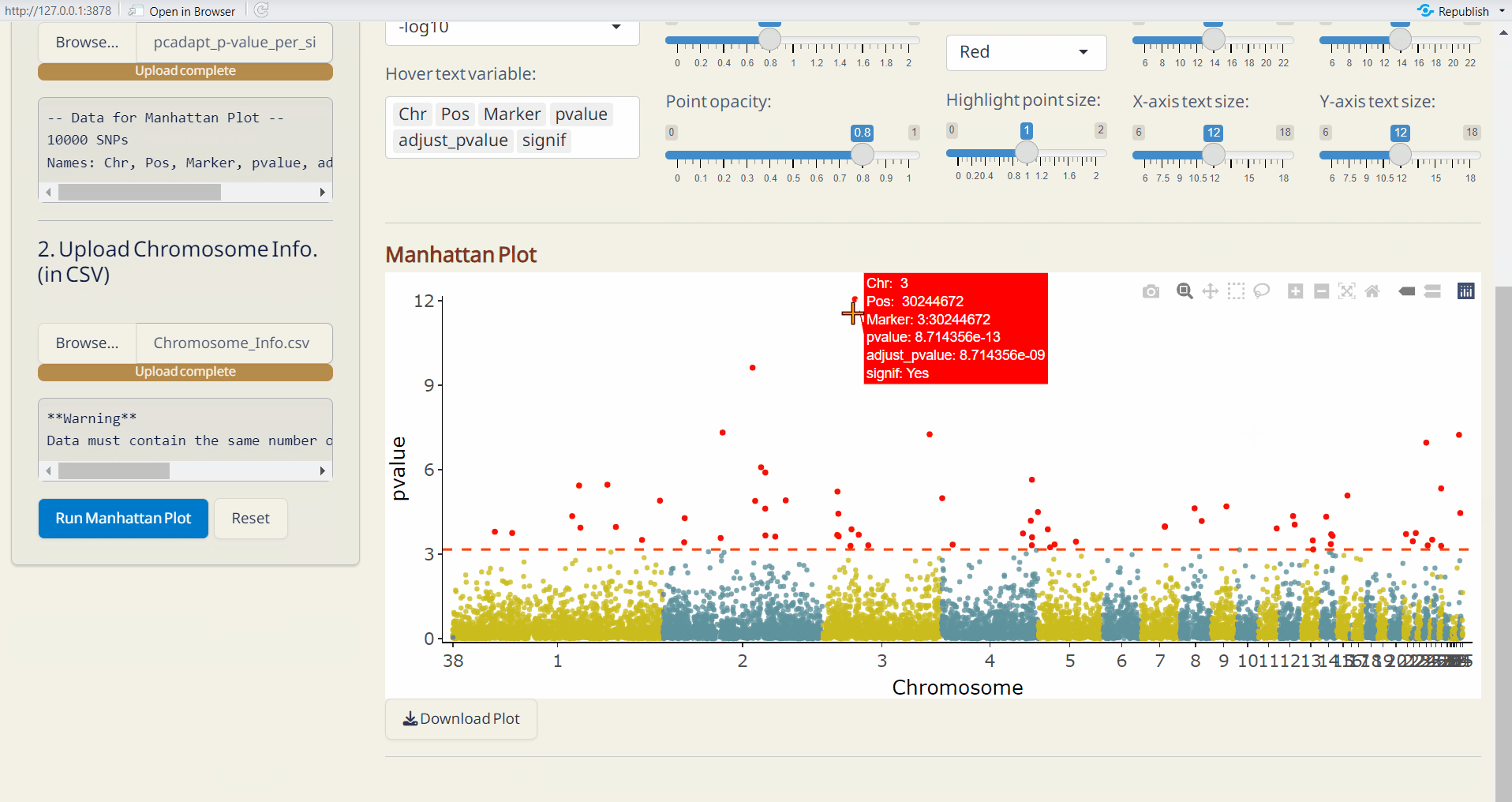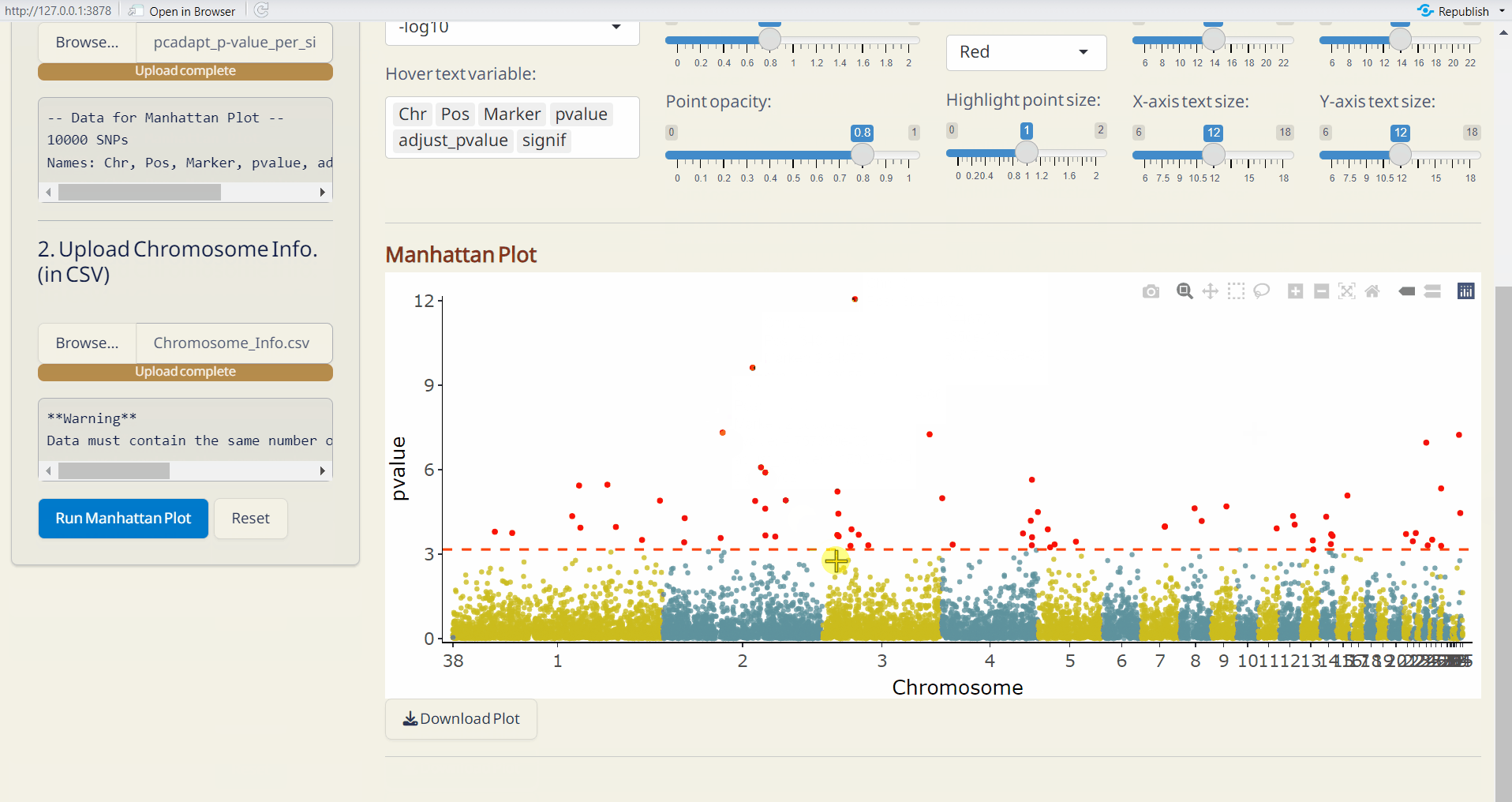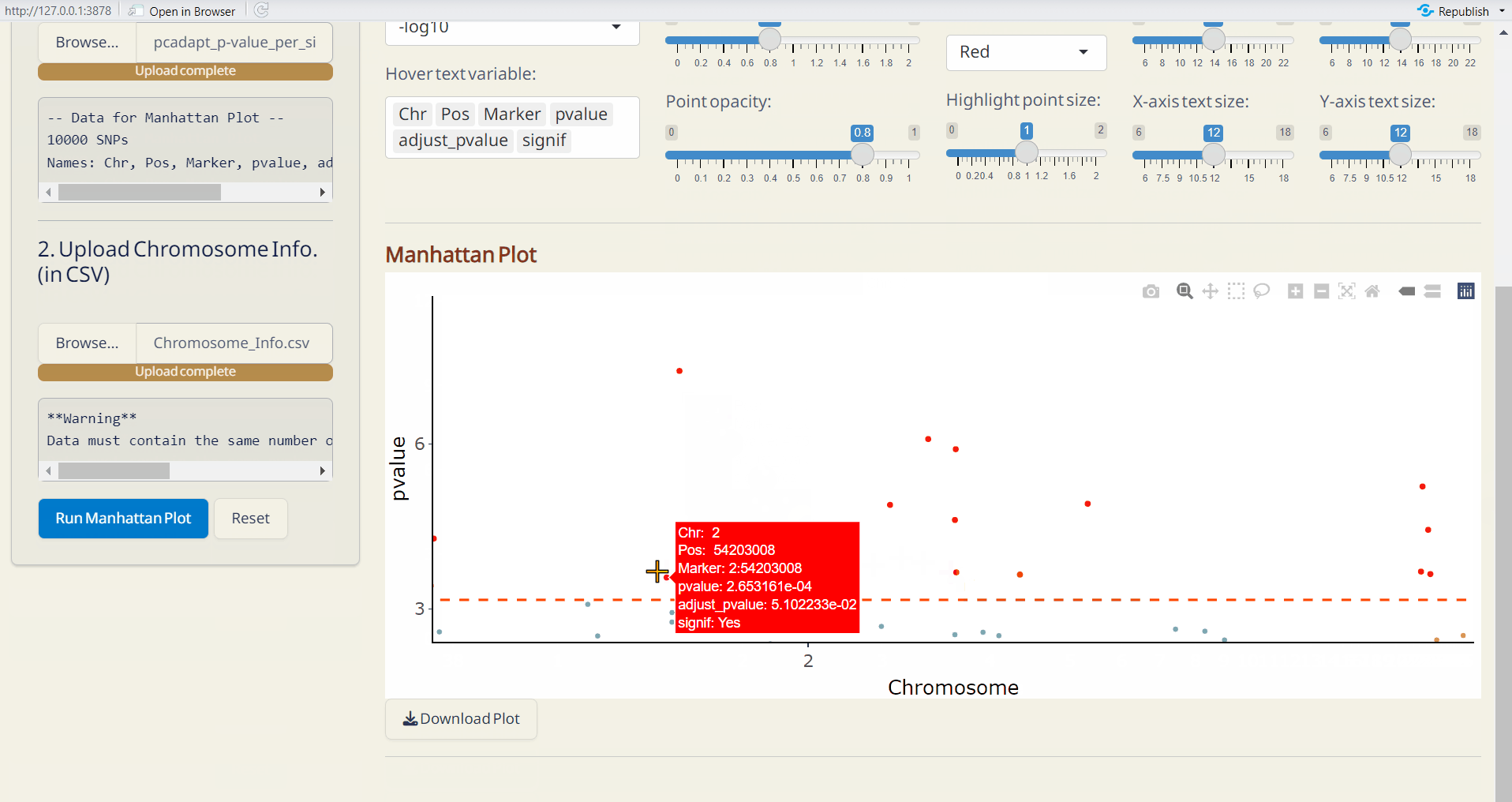
- pcadapt Manhattan Plot (PDF): A Manhattan plot visualizing the p-values per site across the genome. Significant SNPs are highlighted in red.
- pcadapt QQ Plot (PDF): A QQ plot comparing the distribution of observed p-values to the expected distribution under the null hypothesis.
- pcadapt Histogram of p-values (PDF): A histogram showing the distribution of p-values across all sites.
- pcadapt Histogram of Test Statistics (PDF): A histogram showing the distribution of test statistics across all sites.
- pcadapt Significant SNPs (CSV): A table listing SNPs identified as significant by pcadapt, including their site info., p-values, and adjusted p-values.
6.2 OutFLANK
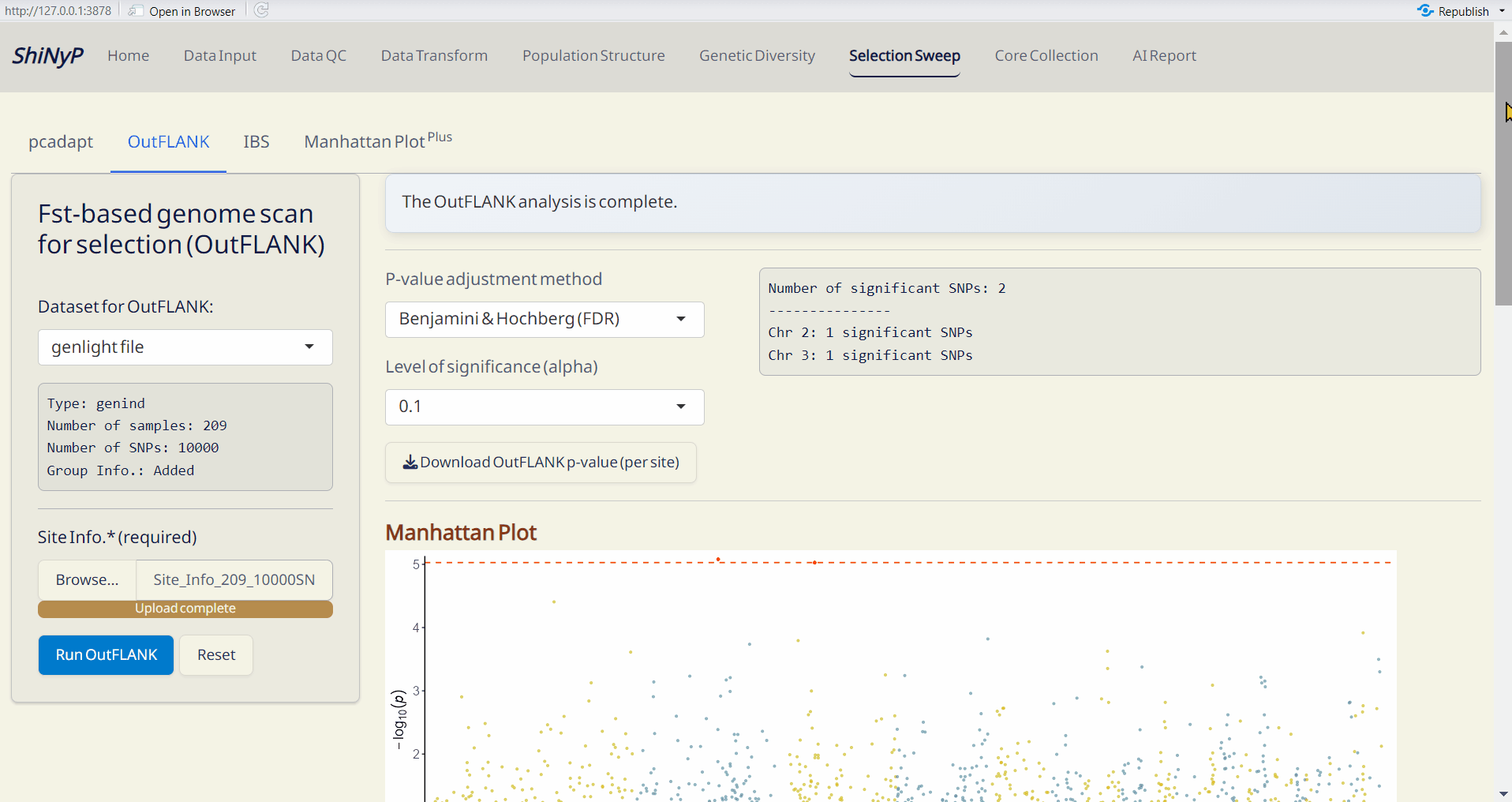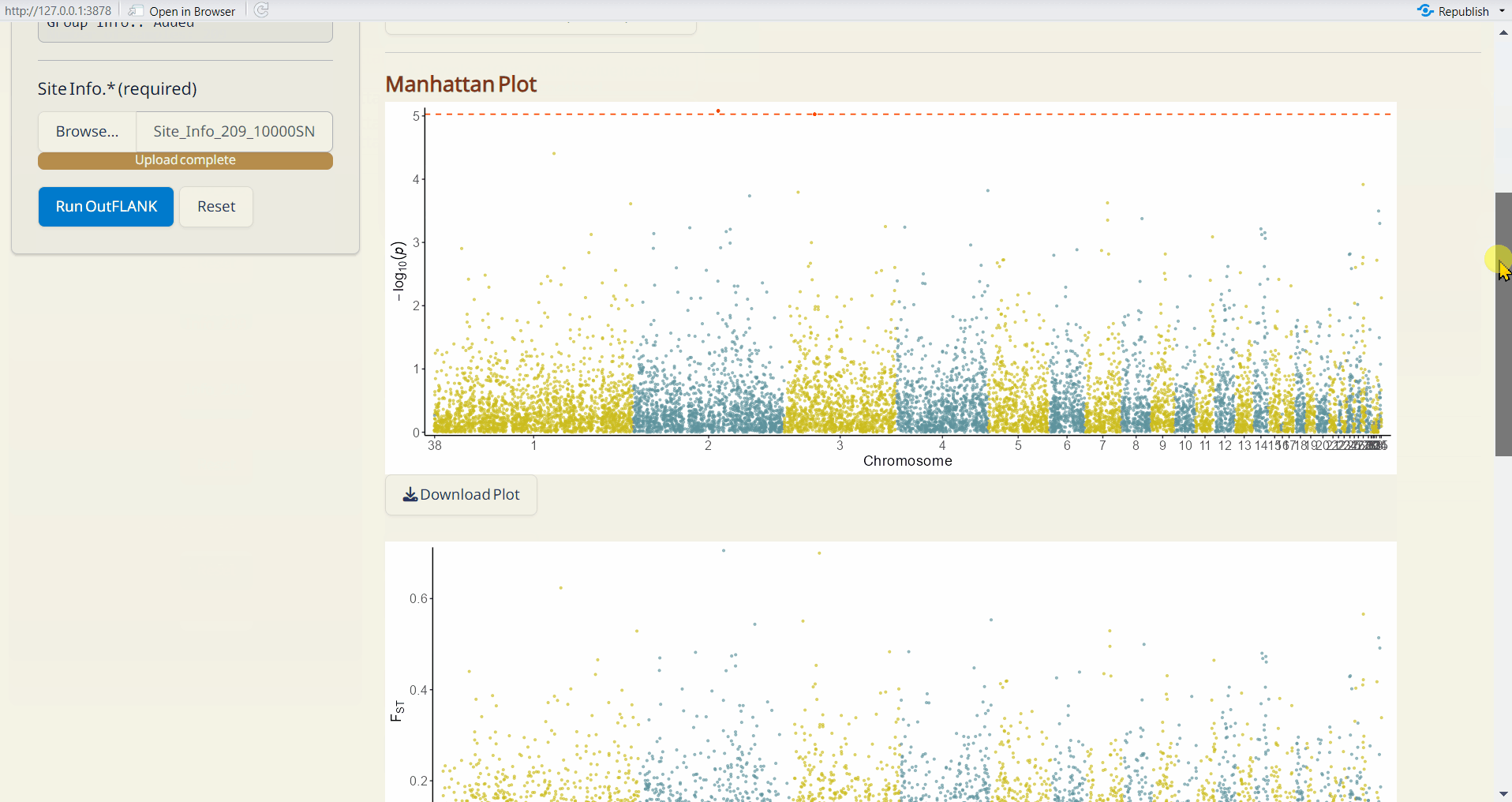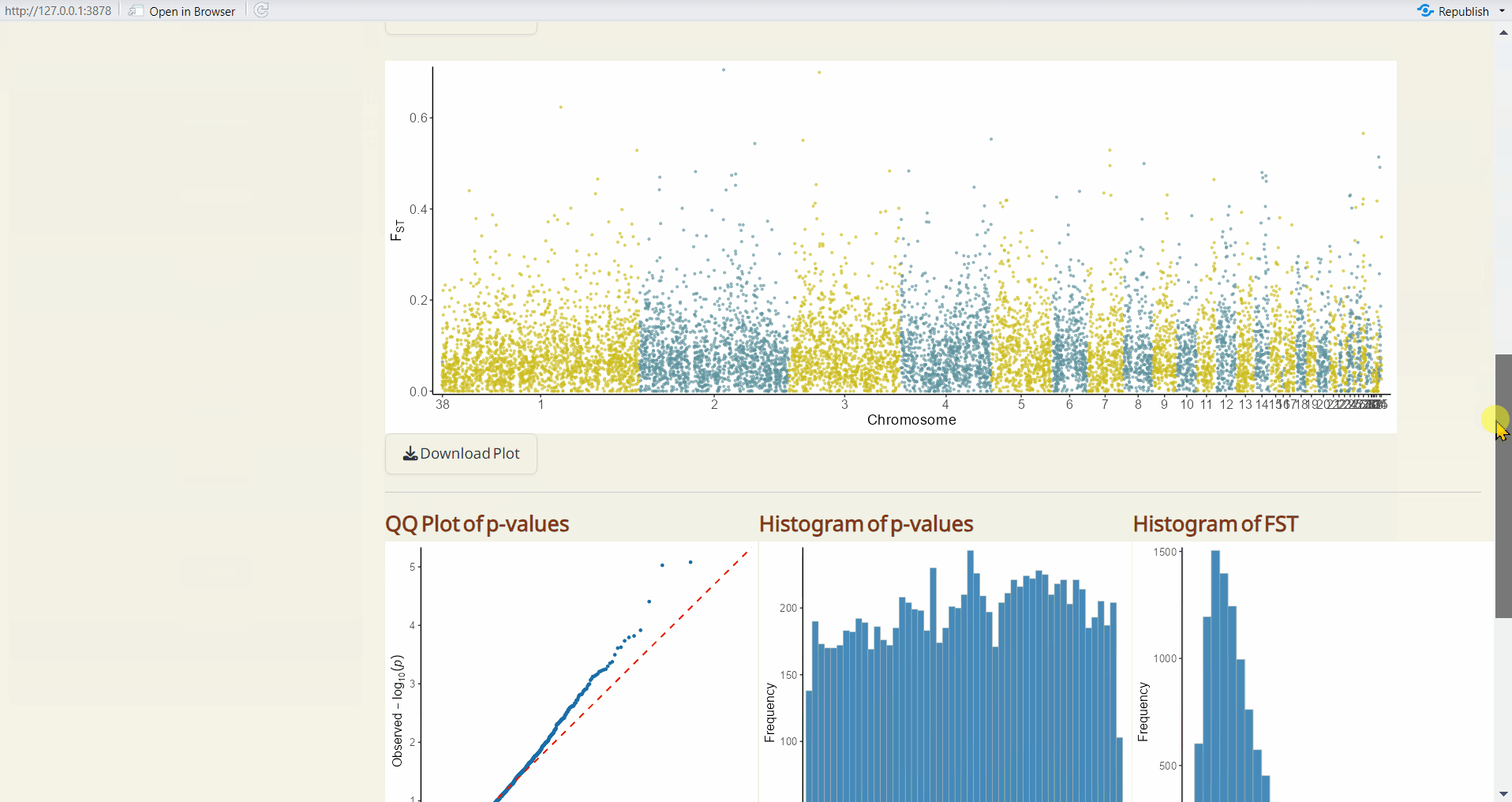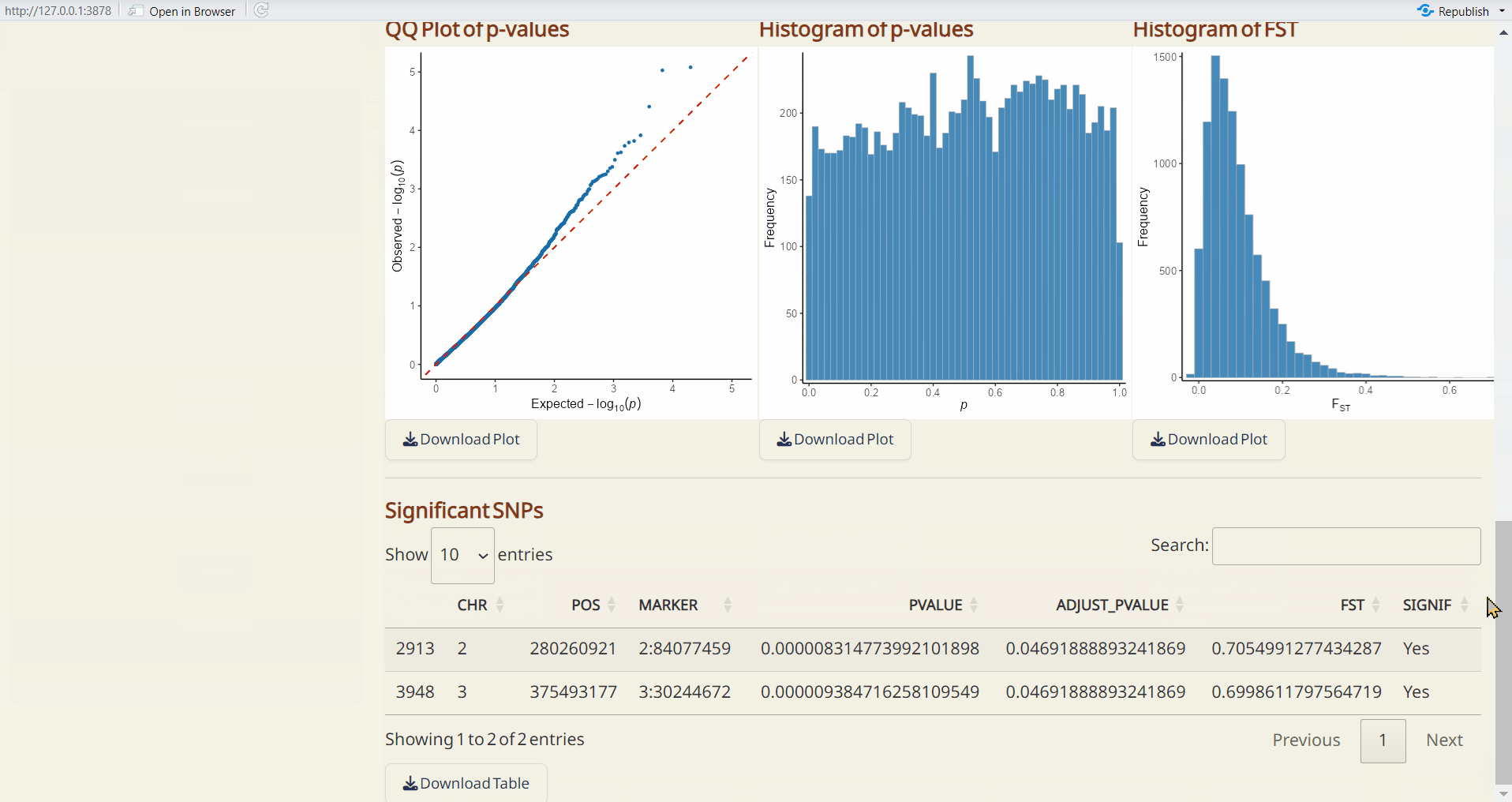
A Fst-based approach detects selection signals by comparing genetic differentiation between defined group assignments (Whitlock and Lotterhos 2015). For more information, visit https://rpubs.com/lotterhos/outflank.
Required Files:
genlight or genind (faster) with ‘Group Info.’, downloadable from Data Transform page after you have Group Info.
Site Info. (RDS) of the current genlight downloadable from Data Input or Data QC pages.
Outputs:
- OutFLANK p-value per site (RDS): A dataset containing p-values and adjusted p-values for each site.
- OutFLANK Manhattan Plot (PDF): A Manhattan plot visualizing the p-values per site across the genome. Significant SNPs are highlighted in red.
- OutFLANK QQ Plot (PDF): A QQ plot comparing the distribution of observed p-values to the expected distribution under the null hypothesis.
- OutFLANK Histogram of p-values (PDF): A histogram showing the distribution of p-values across all sites.
- OutFLANK Histogram of Fst (PDF): A histogram showing the distribution of Fst values across all sites.
- OutFLANK Significant SNPs (CSV): A table listing SNPs identified as significant by OutFLANK, including their site info., Fst values, and p-values.
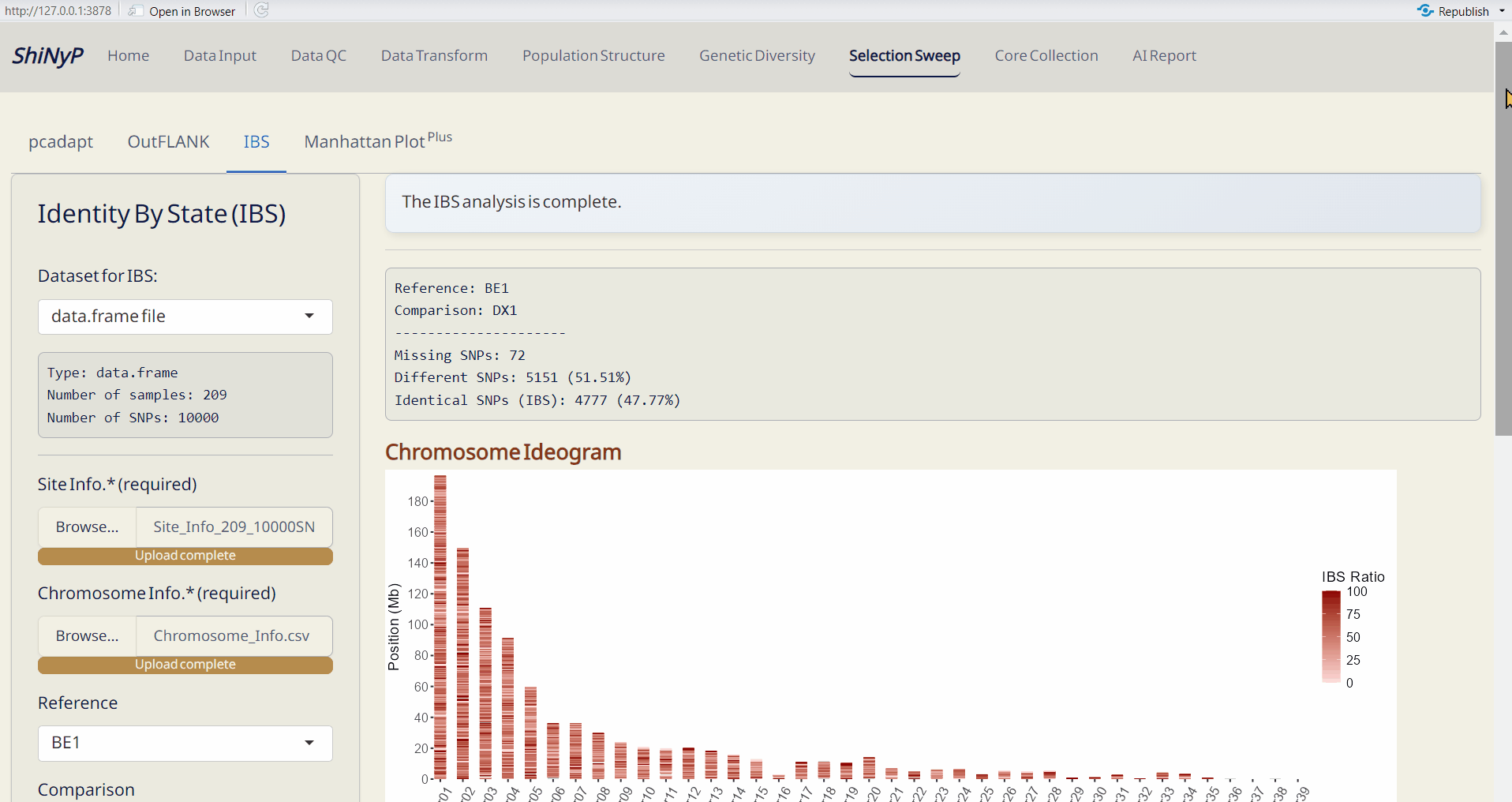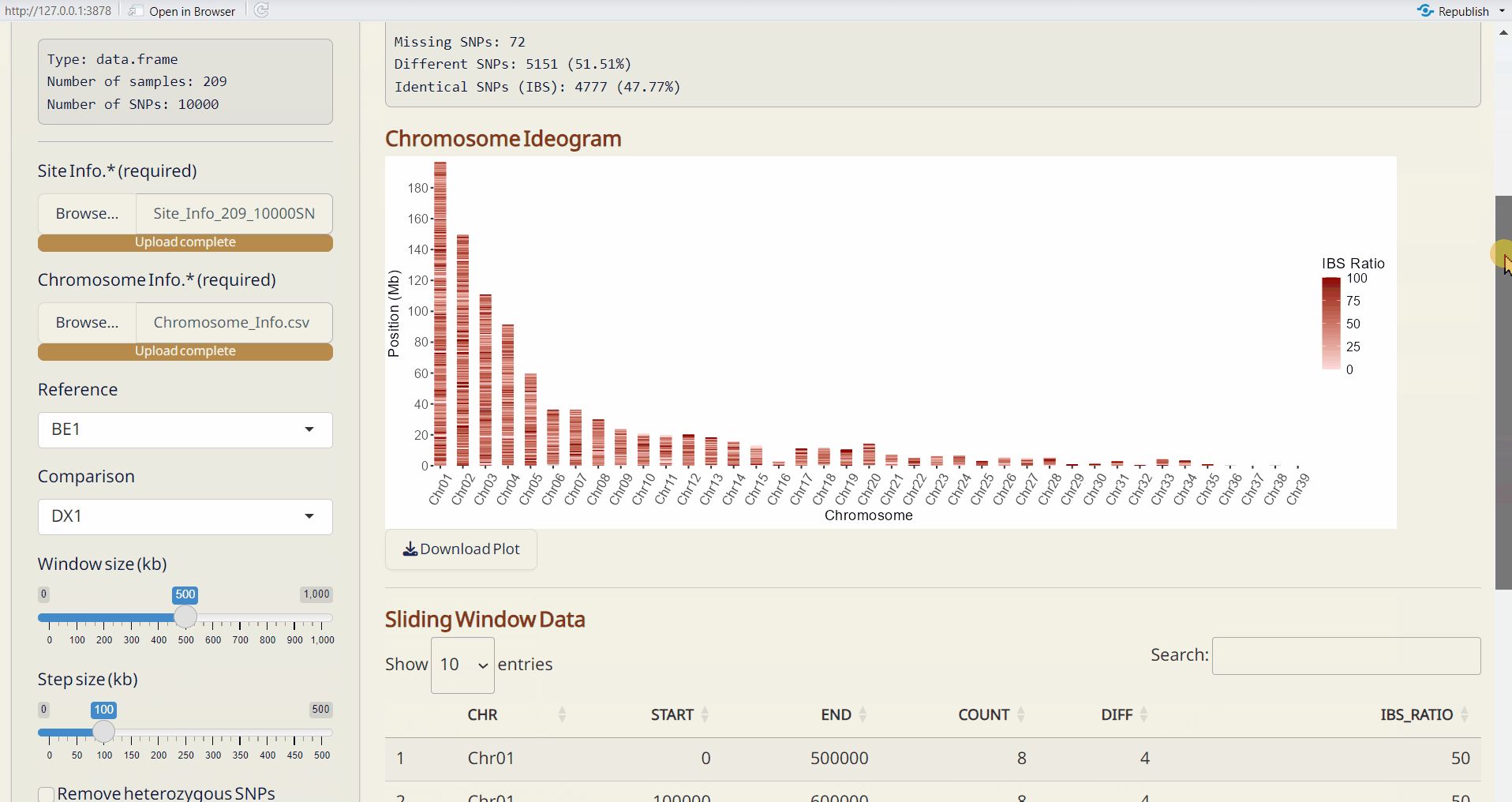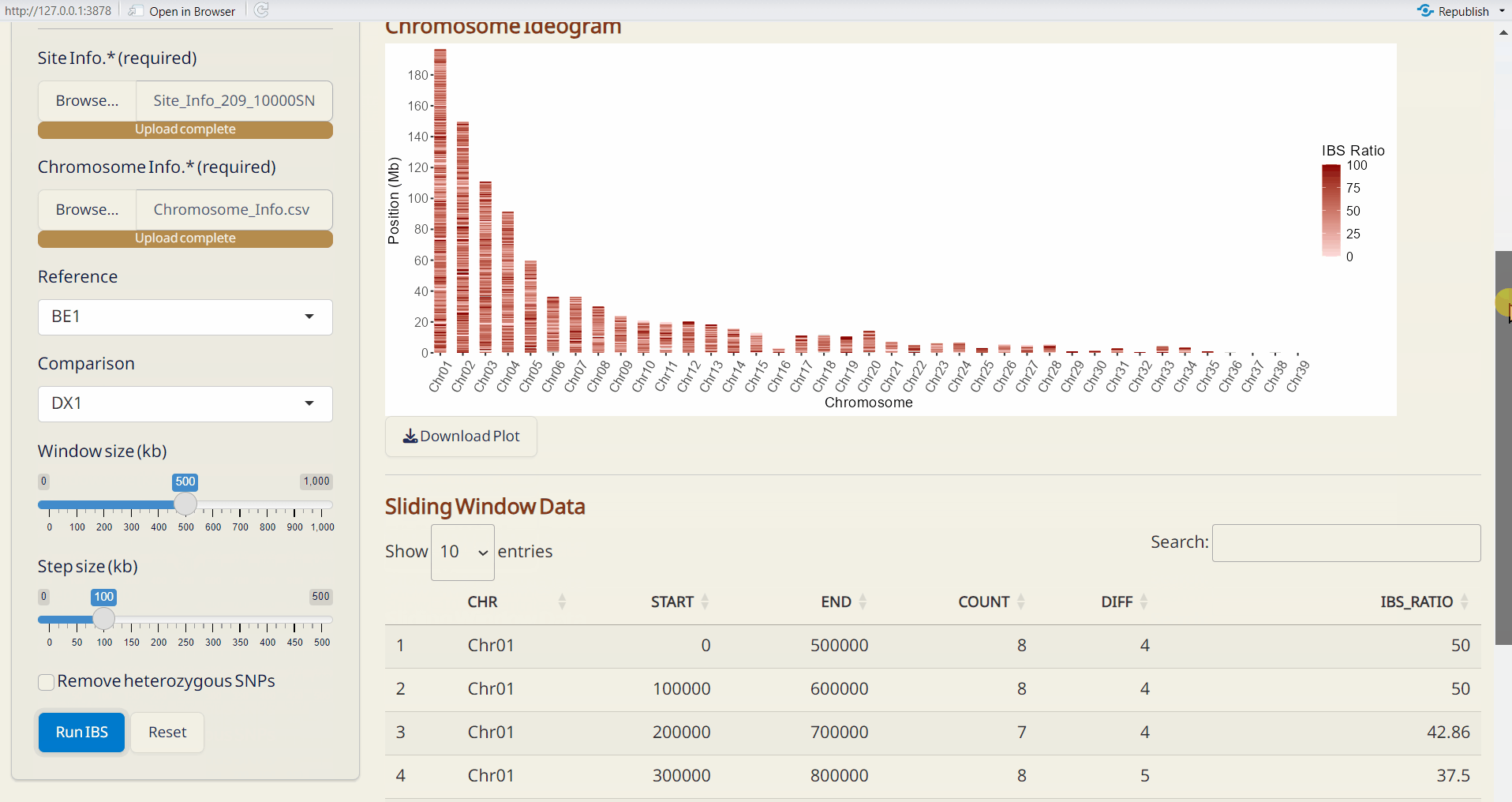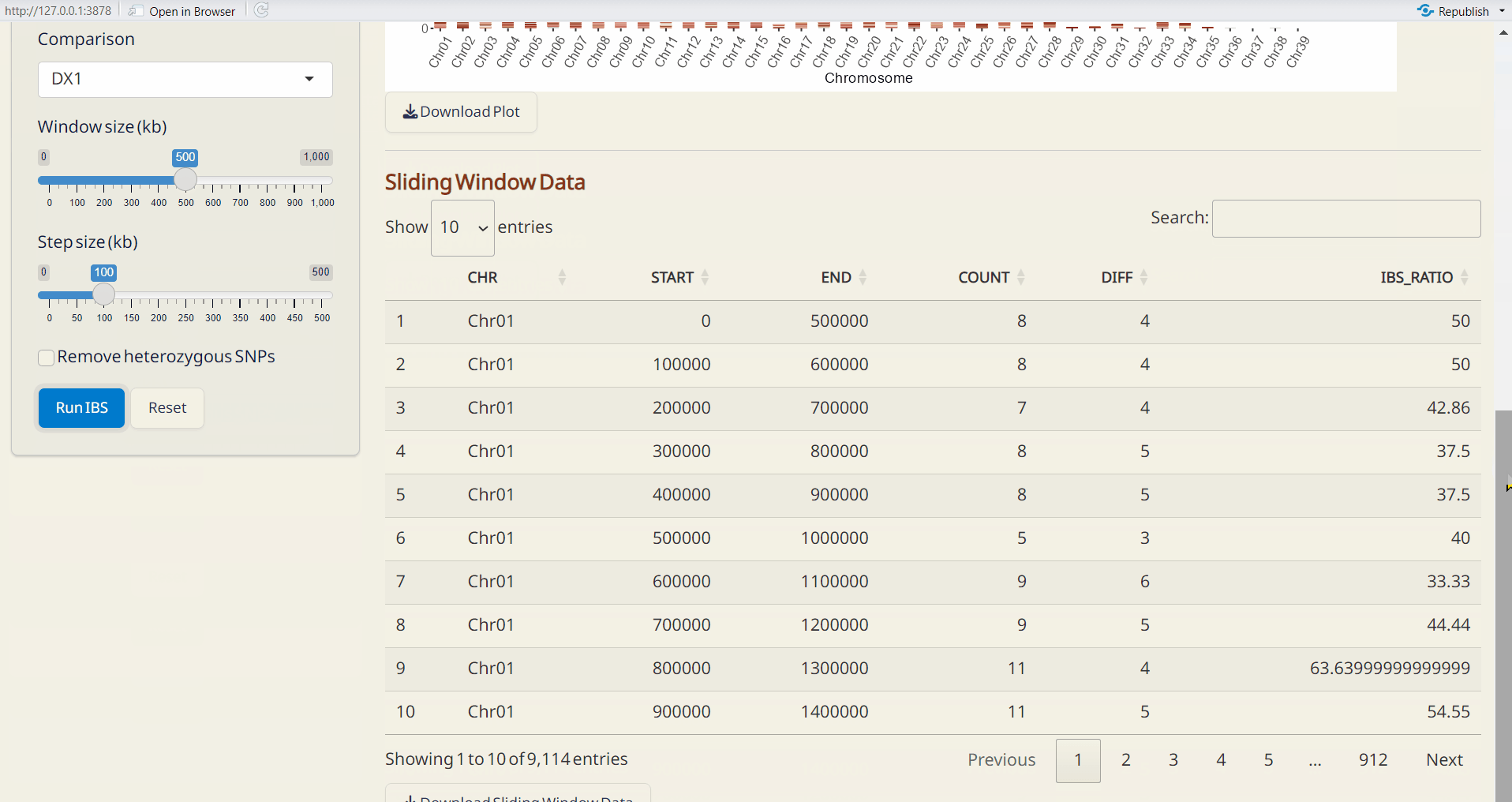
6.3 IBS (Identity By State)
An approach to detect differences in genomic regions between pairs of individuals, useful for identifying pedigree relationships.
Required Files:
data.frame
Site Info. (RDS) of the current data.frame, downloadable from Data Input or Data QC pages.
-
Chromosome Info. (CSV): Reference genome information of the current study.
For more details about this file, refer to Section 2.3 (SNP Density).
Steps:
- Upload Site Info. (required).
- Upload Chromosome Info. (CSV) (required).
- Choose the reference and comparison samples.
- Select window size (kb) and step size (kp).
- To remove heterozygous SNPs from the reference sample, click Remove heterozygous SNPs checkbox (optional).
- Click Run IBS to perform IBS analysis.
Outputs:
- Chromosome Ideogram (PDF): An ideogram visualizing the IBS results, using a gradient palette to represent the differences across chromosomes.
- Sliding Window Data (CSV): A sliding window dataset with IBS results, including SNP count, different SNPs, and the ratio of different SNPs per window.
6.4 Manhattan Plot Plus
Customize your phylogenetic tree plot based on the results from Genetic Diversity/Diversity Parameter, Selection Sweep/pcadapt, or Selection Sweep/OutFLANK.
Required Files:
Genetic Diversity per Site (Genetic_Diversity_per_Site.rds), pcadapt p-value per Site (pcadapt_p-value_per_site.rds), or OutFLANK p-value per Site (OutFLANK_p-value_per_site.rds).
-
Chromosome Info. (CSV): Reference genome information of the current study.
For more details about this file, refer to Section 2.3 (SNP Density).