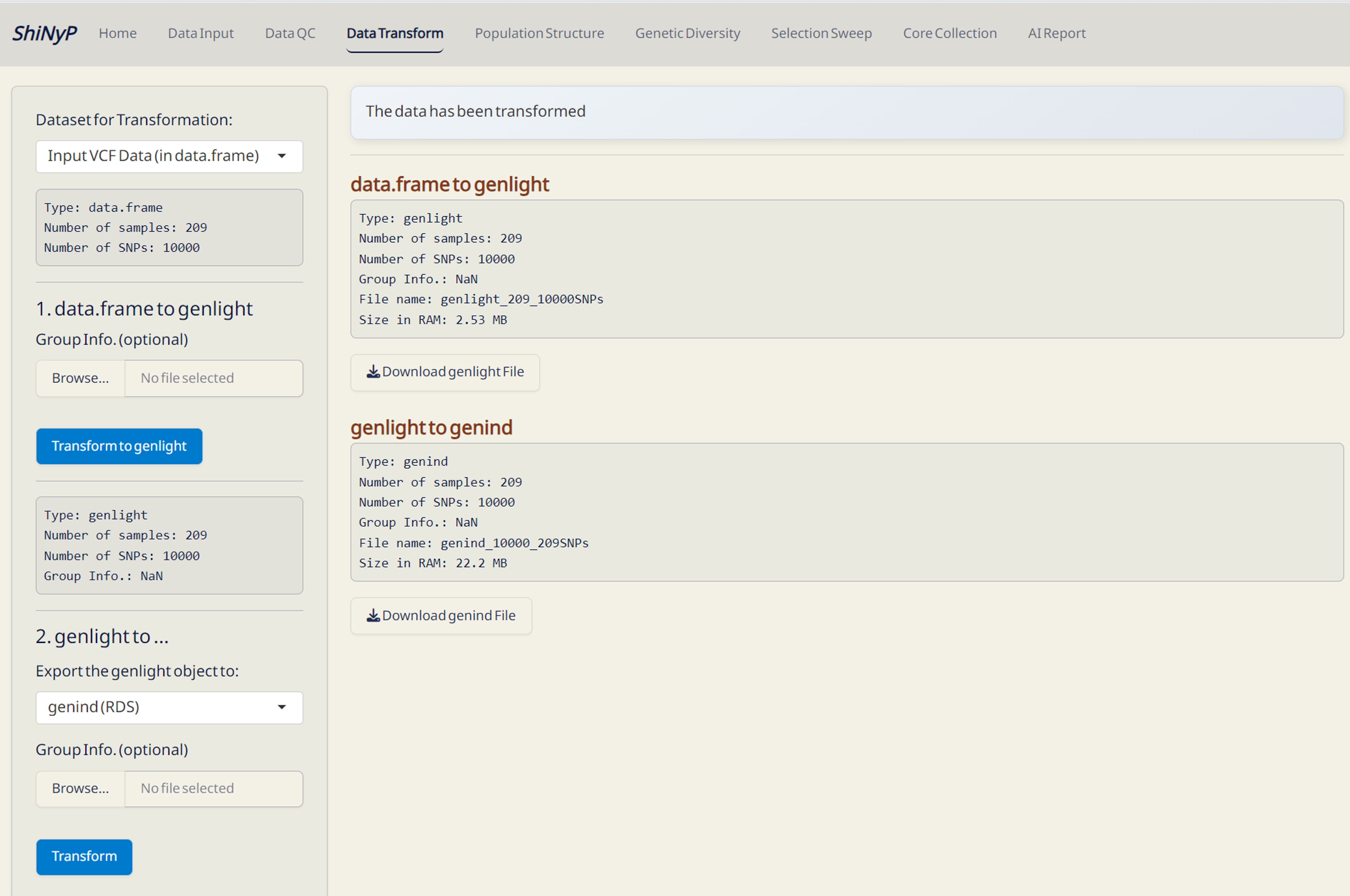
3 Data Transform
➡️ This section allow you to convert your SNP data in data.frame into multiple formats, including genind, genind and PLINK.
Required File:
- Input VCF Data (data.frame file) from the Data Input page or
- Post-QC Data (data.frame file) from the Data QC page.
Step 1: Transform data.frame to genlight
- Click Transform to genlight.
- Download the generated genlight (in RDS format) to skip VCF or data.frame upload next time by directly importing this file.
Step 2: Transform genlight to others
Select the desired data format to export from genlight and click Transform.
Outputs:
genlight (RDS): genlight file with Group Info. — required for downstream analysis.
-
genind (RDS): One of the input format for ShiNyP DAPC subpage, optimized for DAPC analysis to reduce computation time.
Download an example of Group Info. (CSV).
This file should have at least two columns: “ID” and “Group”.
Note: You can obtain a template after completing a DAPC analysis (Section 4.2) first to generate the initial file (“DAPC_Group_Info.csv”). If your SNP data is large, you can create and use a core SNP set as input to obtain DAPC results more efficiently. You can then expand this file based on your own metadata (e.g., origin, type).
The following transformed files will be generated at the specified path you provide.
- PLINK (PED & MAP): Input format for PLINK program, designed to perform a range of basic and large-scale SNP analyses.
- GenAlEx (CSV): Input format for GenAlEx program, offers a wide range of population genetic analysis in Excel.
- LEA (GENO & LFMM): Input format for LEA R package, designed for population genomics, landscape genomics and genotype-environment association tests.
- GDS (GDS): Input format for SNPRelate R package, designed for efficient SNP data analysis.
- STRUCTURE (STR): Input format for STRUCTURE program, used for inferring population structure.
- fastStructure (STR): Input format for fastStructure program, used for inferring population structure from large SNP data.
- PHYLIP (TXT): Input format for PHYLIP program, used for phylogenetic tree reconstruction and evolutionary analysis.
- Treemix (GZ): Input format for Treemix program, designed for modeling population splits and migration events.
- BayeScan (TXT): Input format for BayeScan, used for detecting loci under selection.