2 Data QC
2.1 Sample QC
Required File:
- data.frame file from the Data Input page or
- SNP post-QC data.frame file from the Data QC/SNP QC subpage .
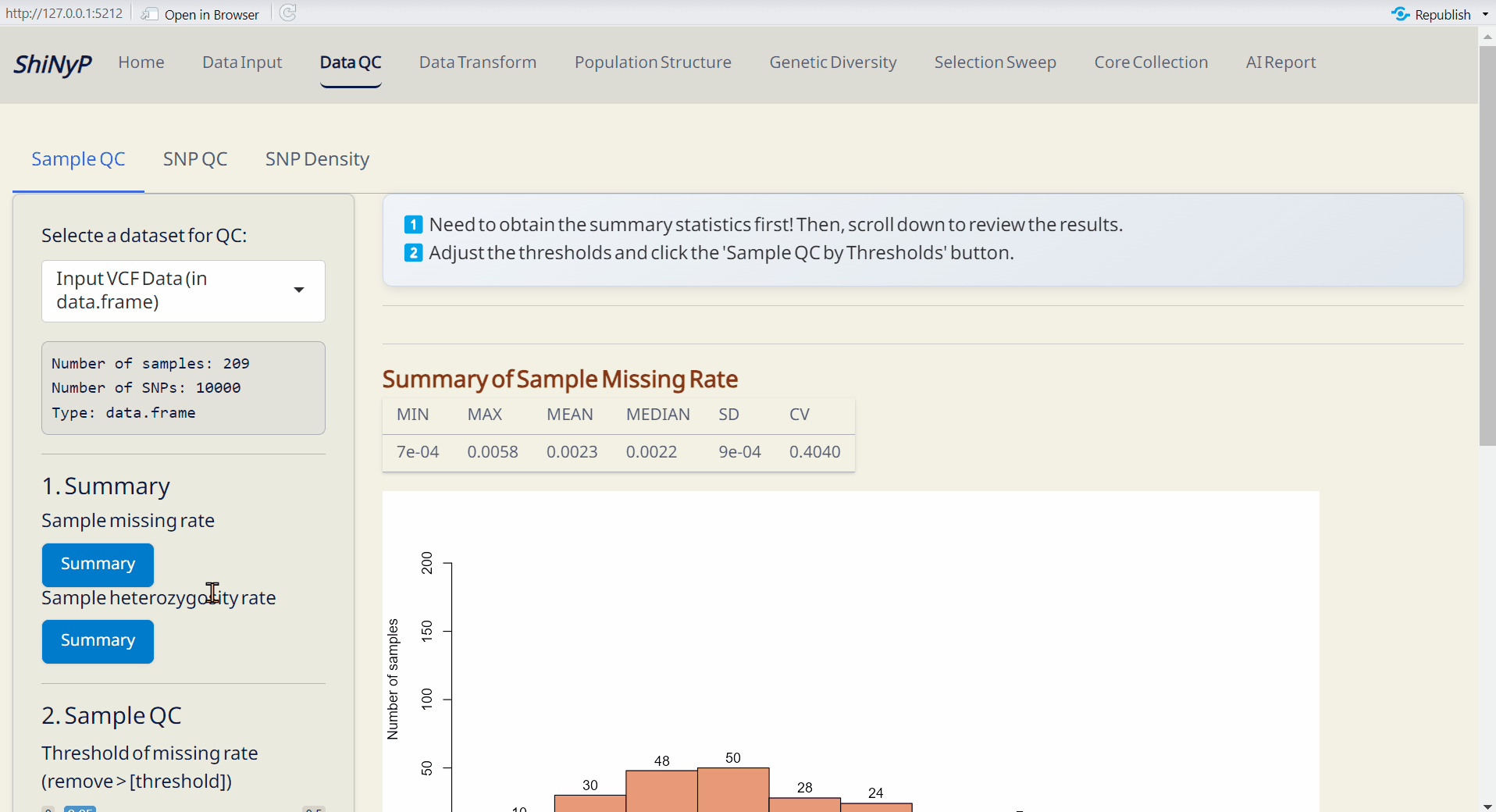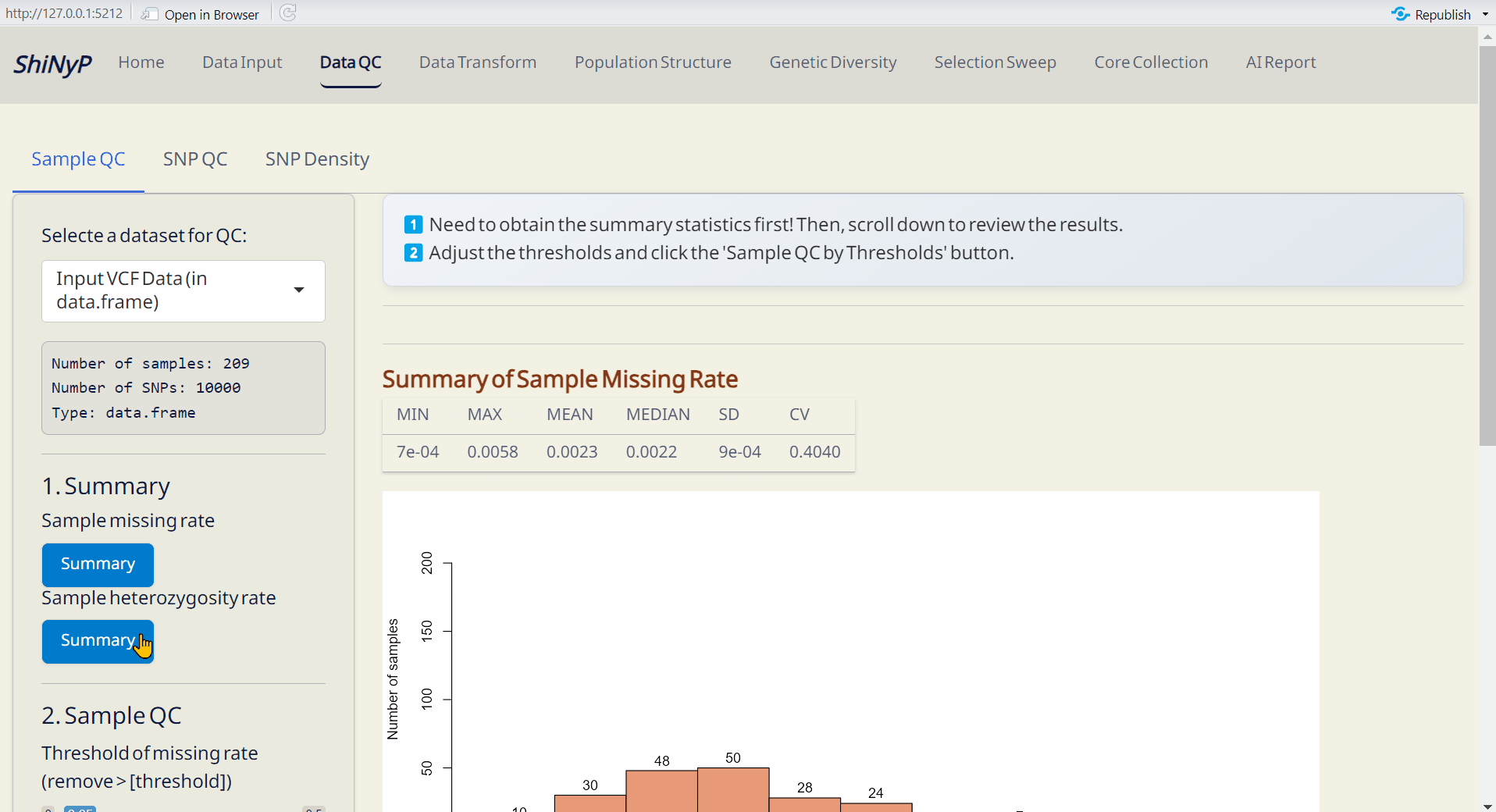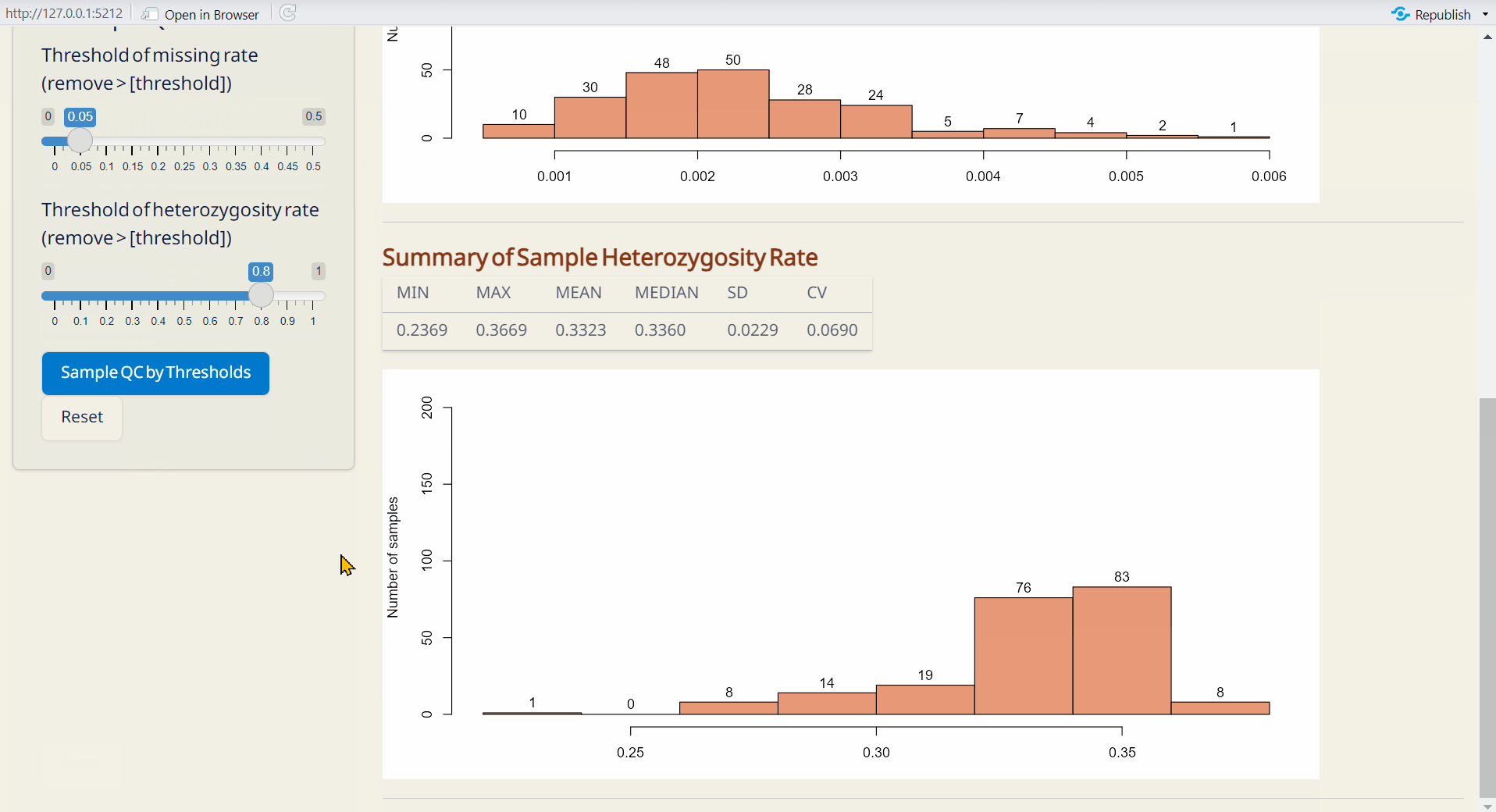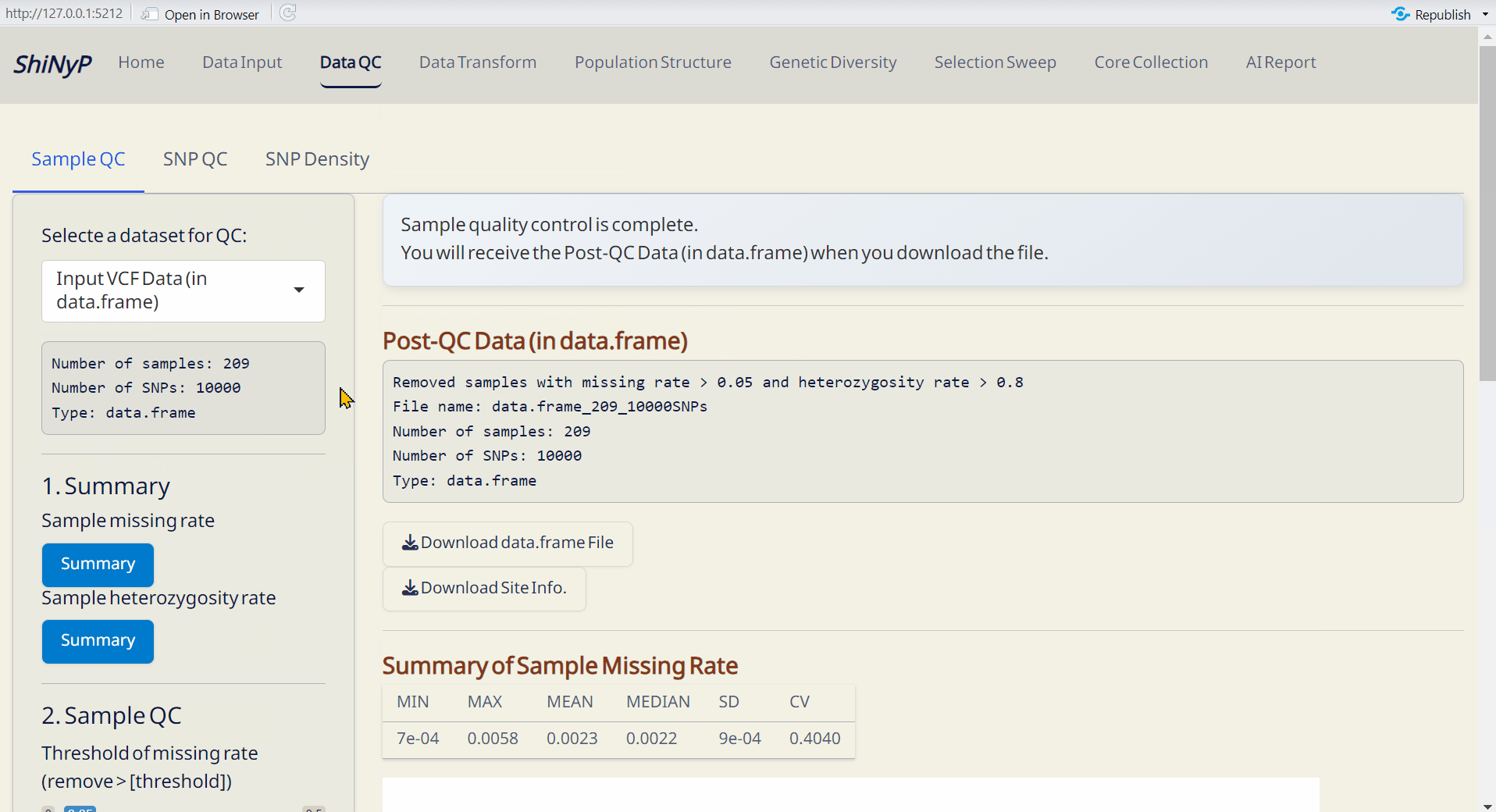
Step 1: Get Summary
First, obtain the sample summary statistics (missing rate and heterozygosity rate) by clicking both Summary buttons and you will see the results.
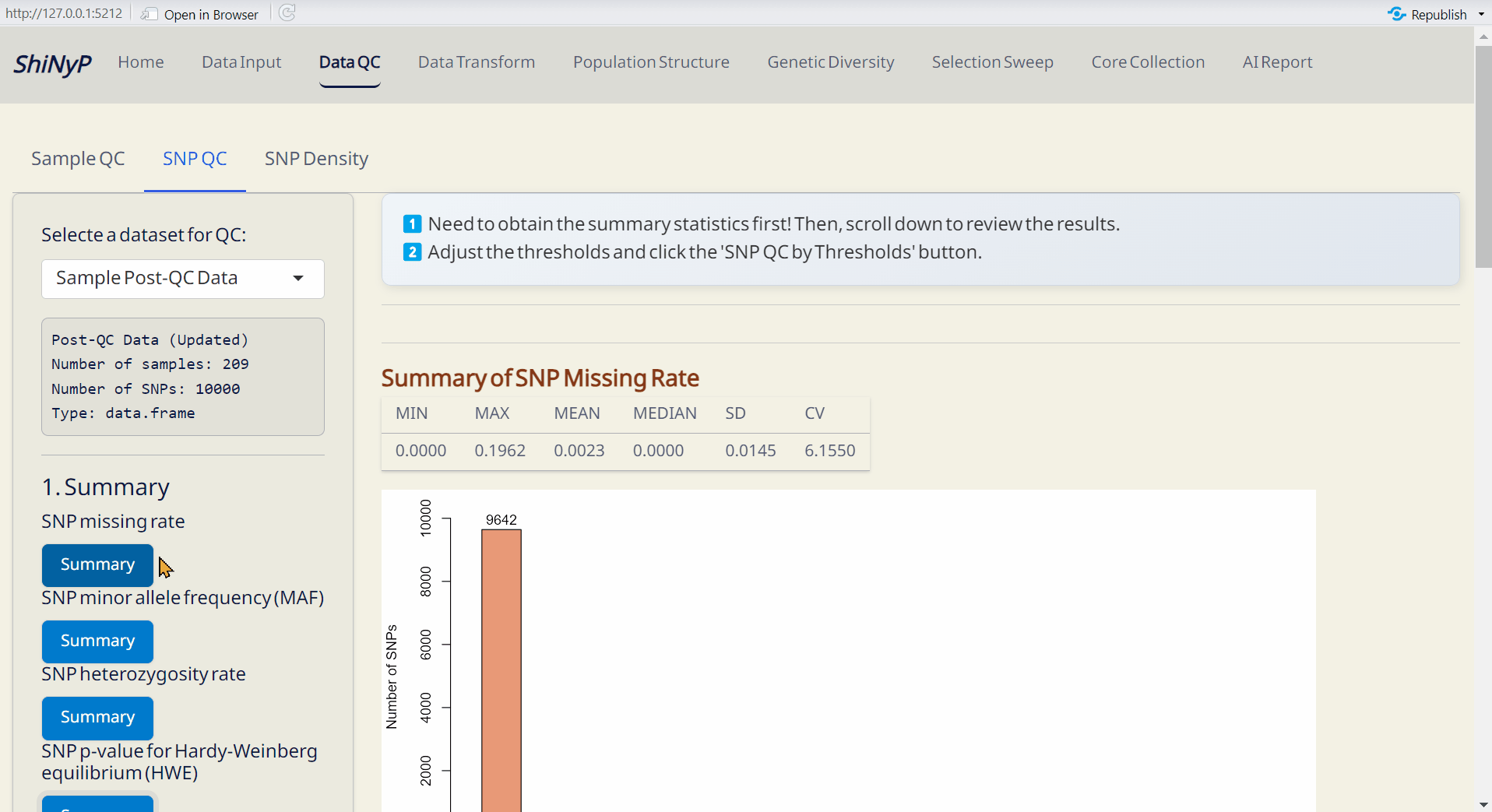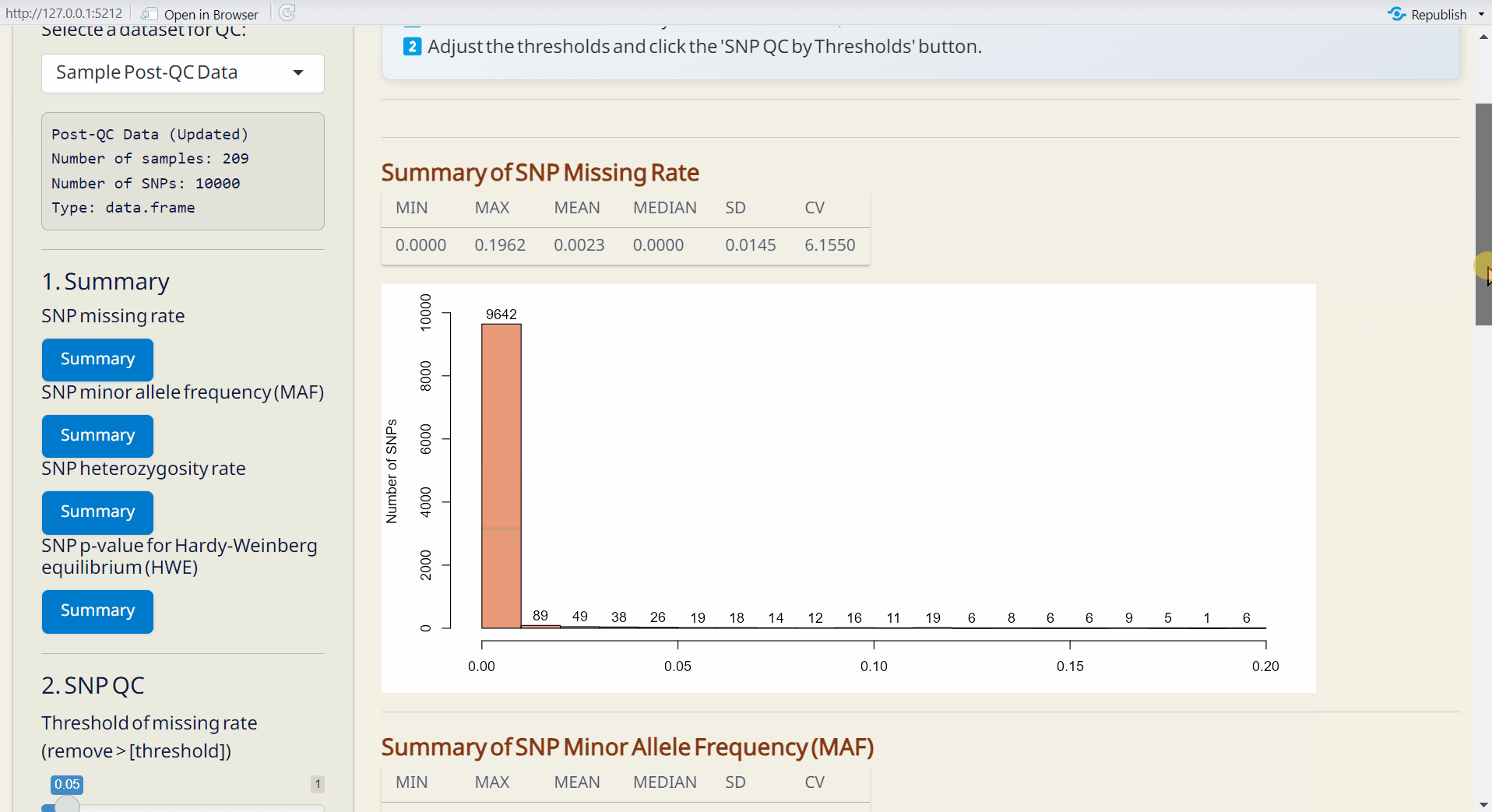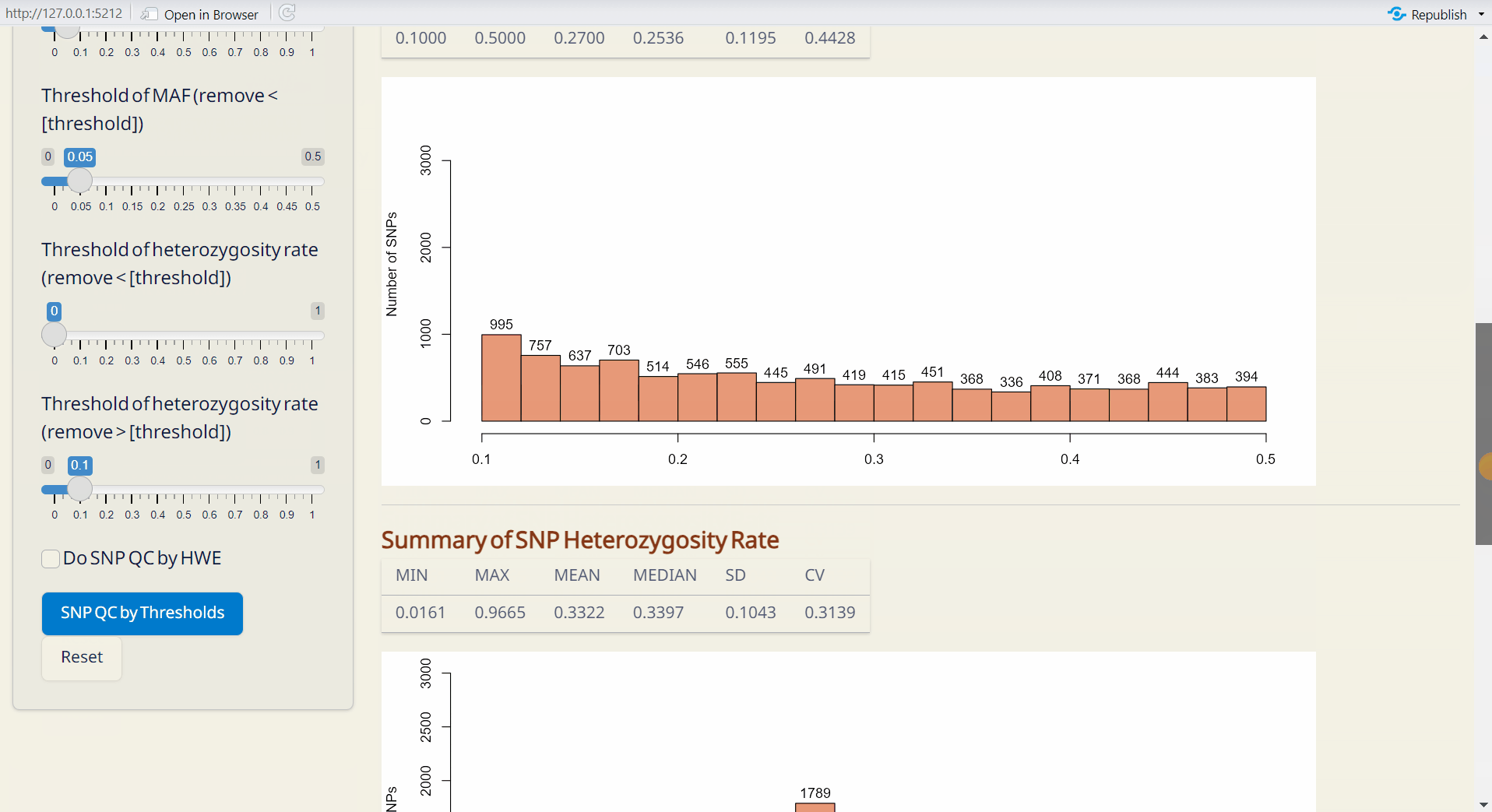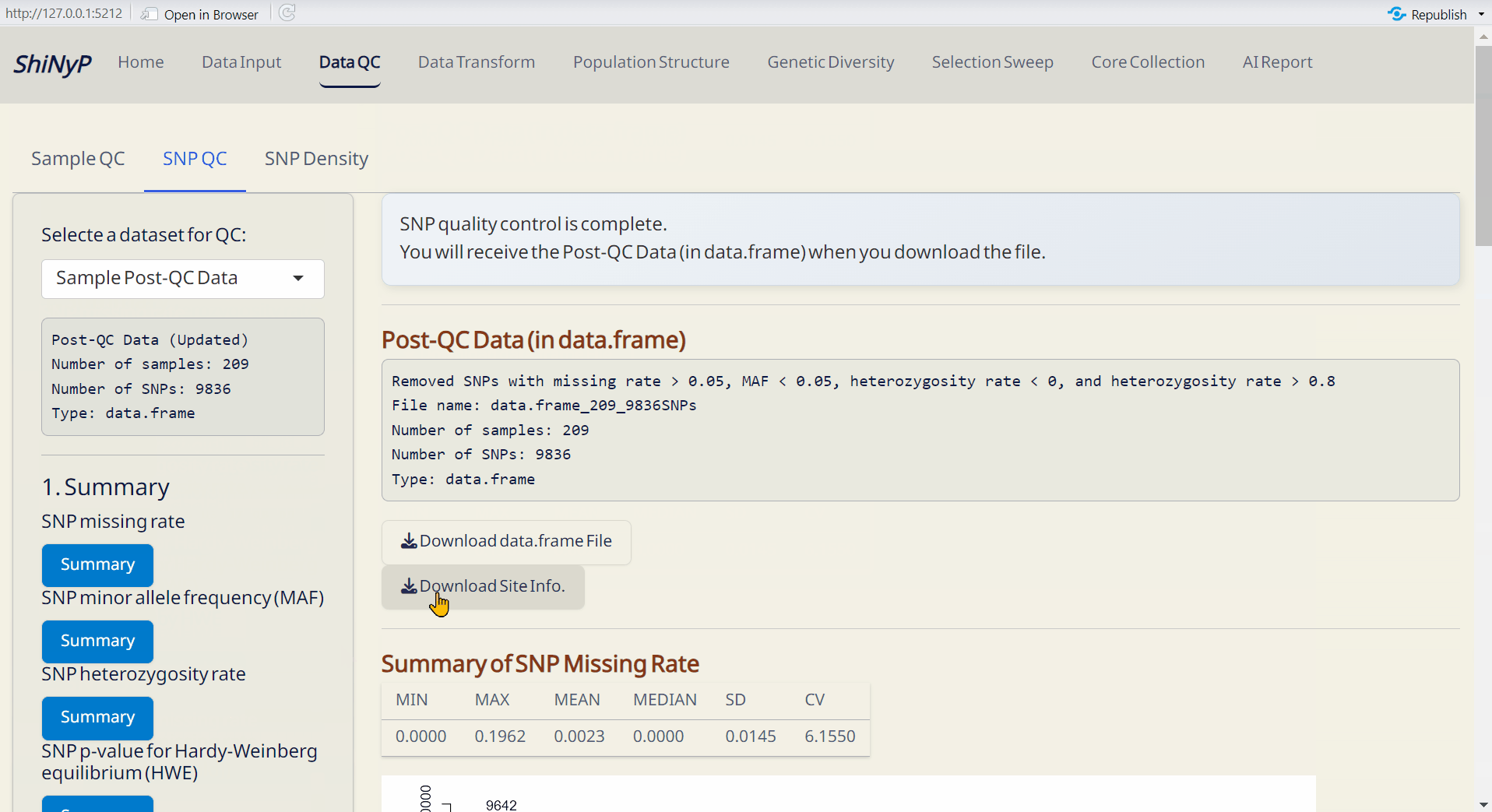
2.2 SNP QC
Required File:
- data.frame file from the Data Input page or
- Sample post-QC data.frame file from the Data QC/Sample QC subpage.
Step 1: Get Summary
First, obtain the SNP summary statistics [missing rate, minor allele frequency (MAF), heterozygosity rate, and Hardy-Weinberg equilibrium (HWE)] by clicking all Summary buttons and you will see the results.
Step 2: Sample QC
Adjust the thresholds and click SNP QC by Thresholds. This will generate the Post-QC data.frame file.
Note: If you prefer not to perform QC based on SNP missing rate or heterozygosity rate, set the missing rate threshold to 1, the MAF to 0, and the heterozygosity rate to 0 and 1. Also, leave the ‘Do SNP QC by HWE’ checkbox unticked to skip QC based on SNP HWE.
2.3 SNP Density
Required Files:
Site Info. (RDS) of the current data.frame, downloadable from Data Input or Data QC pages.
-
Chromosome Info. (CSV): Reference genome information of the current study.
Download an example of Chromosome Info. (CSV).
This file should contain three columns: “Chr”, “Start”, and “End”.
- “Chr” column should specify the chromosome names (as characters, e.g., “Chr01”, “Chr11”)
- “End” column should indicate the length of each chromosome (numeric)
- “Start” column can be set to 0 or 1 for each chromosome.
Steps:
- Upload Site Info. (RDS) and Chromosome Info. (CSV).
- Choose a window size in kilobases (kb).
- Click Summary.
Outputs:
- SNP Density Plot (PDF): An ideogram visualizing SNP density across the genome within a defined window size. A gradient color palette is used to represent varying SNP densities: green for lower densities, yellow for medium densities, and red for higher densities, with grey indicating regions with zero SNP.
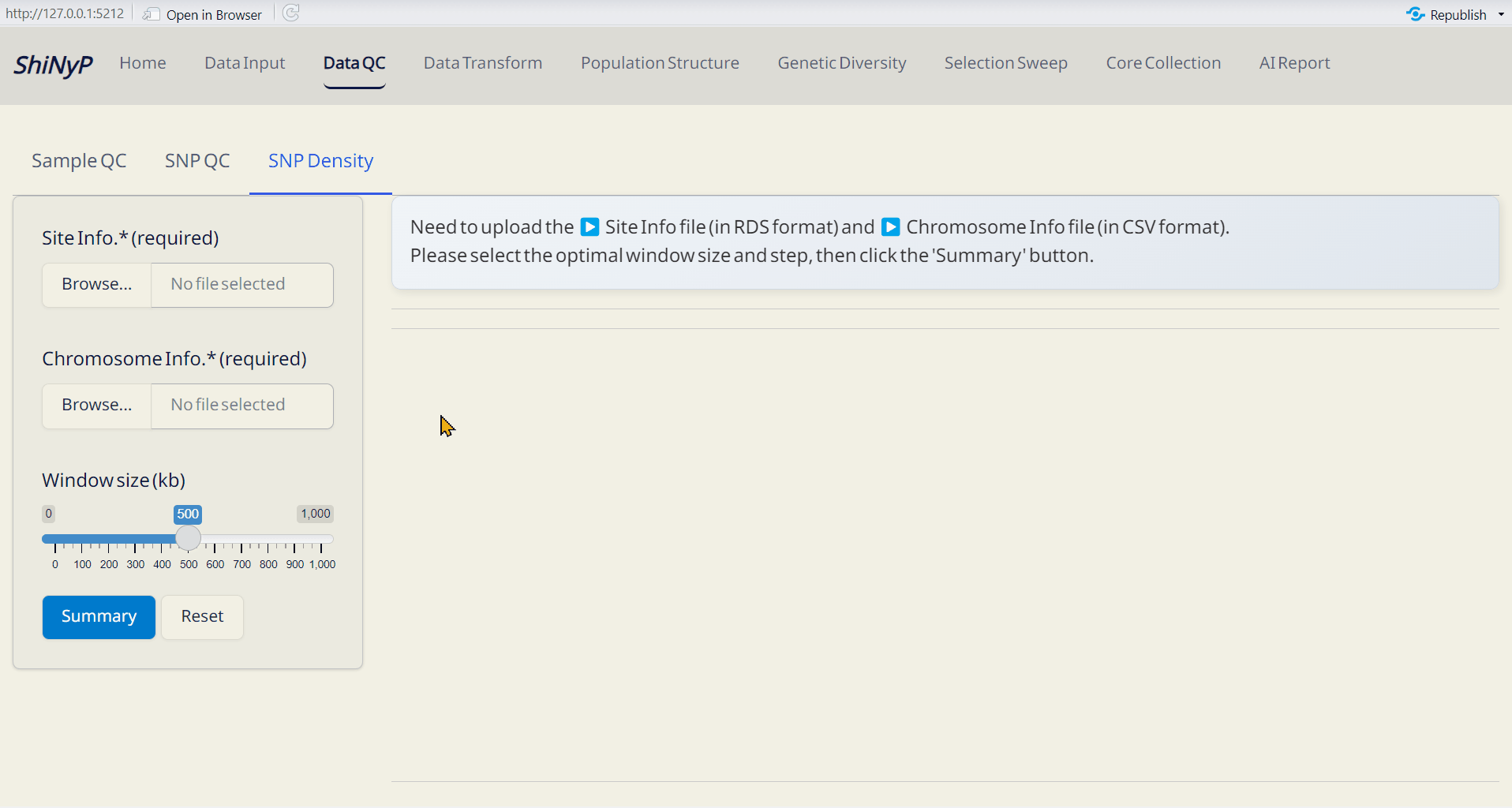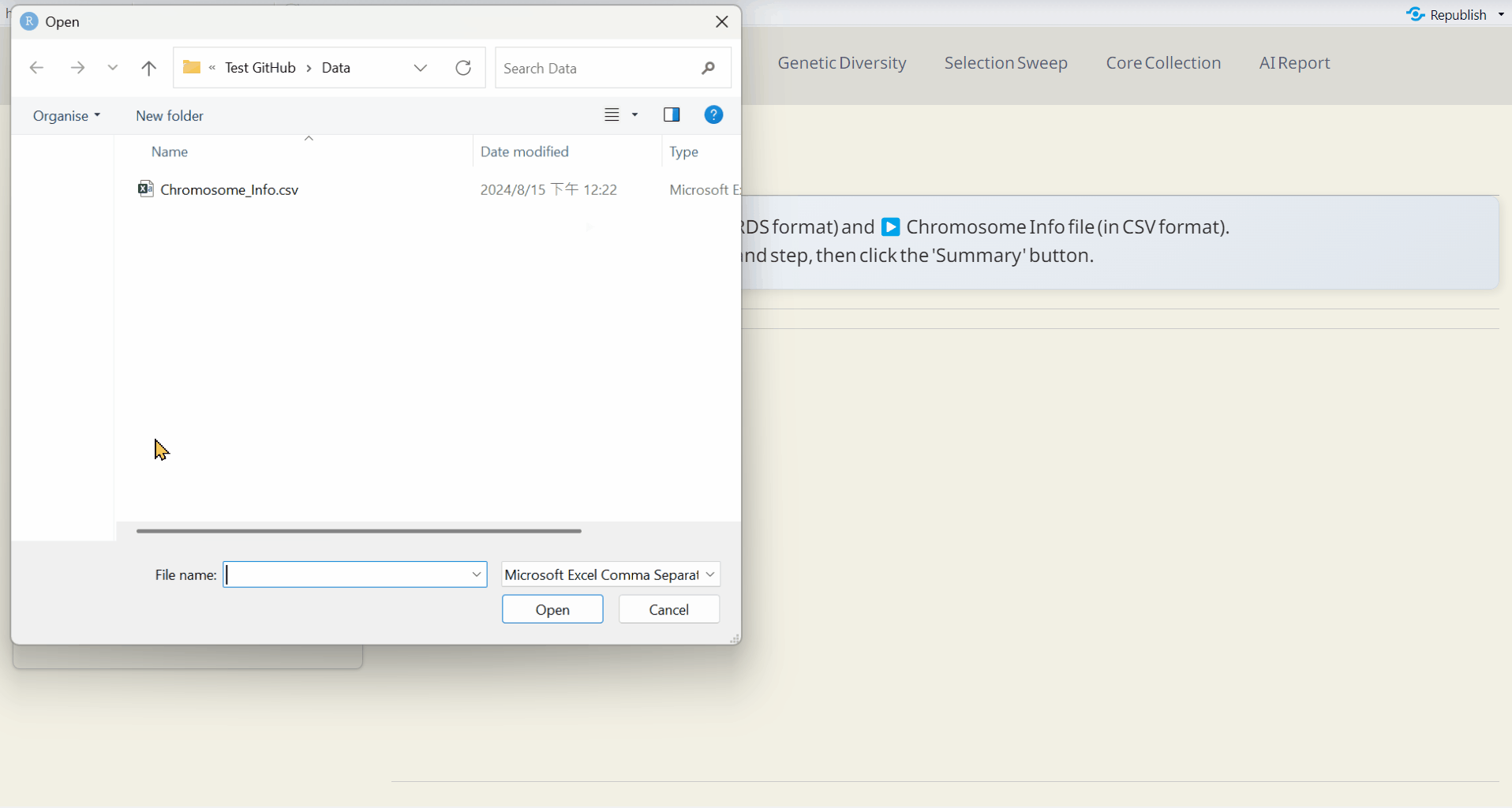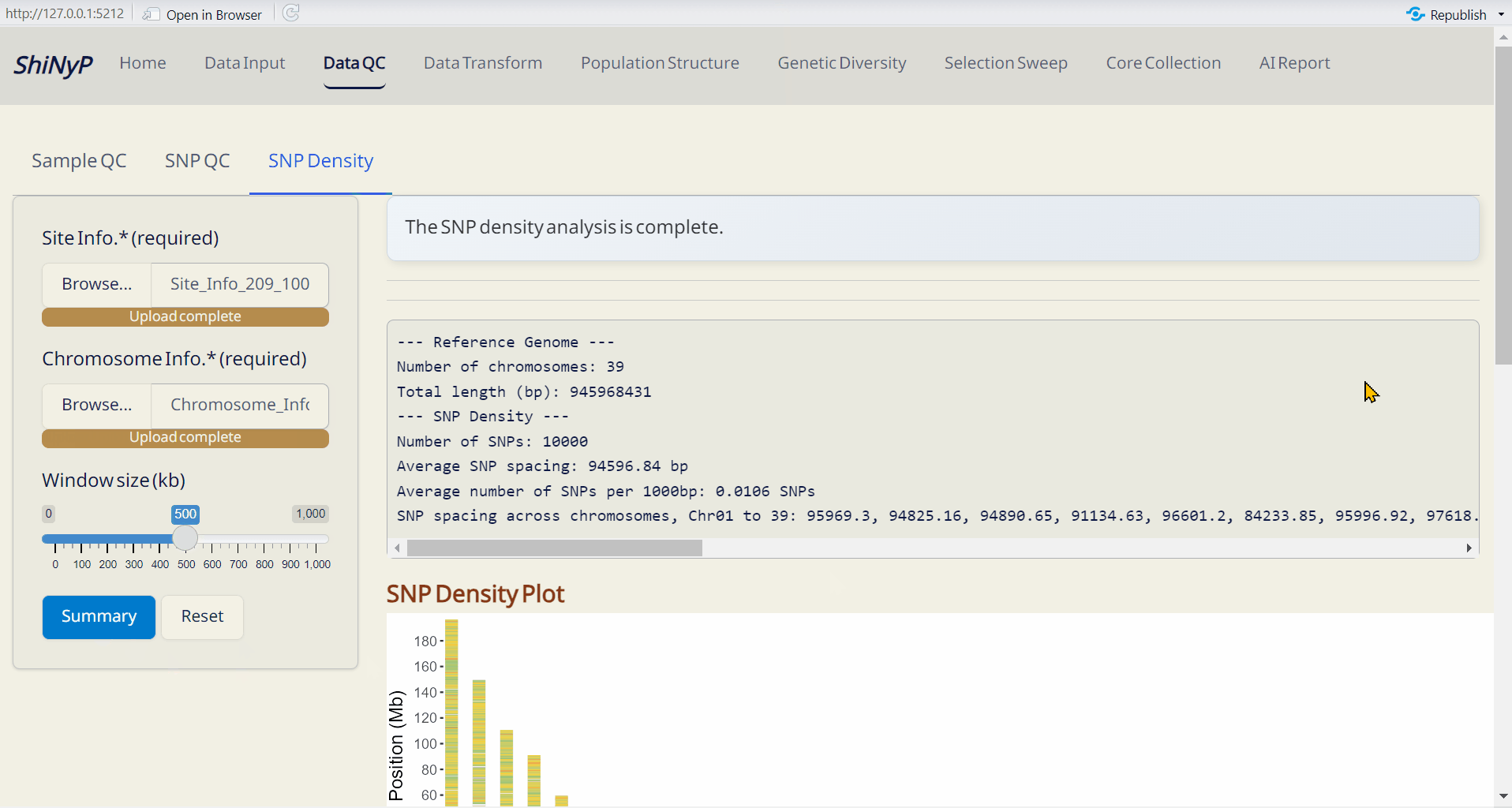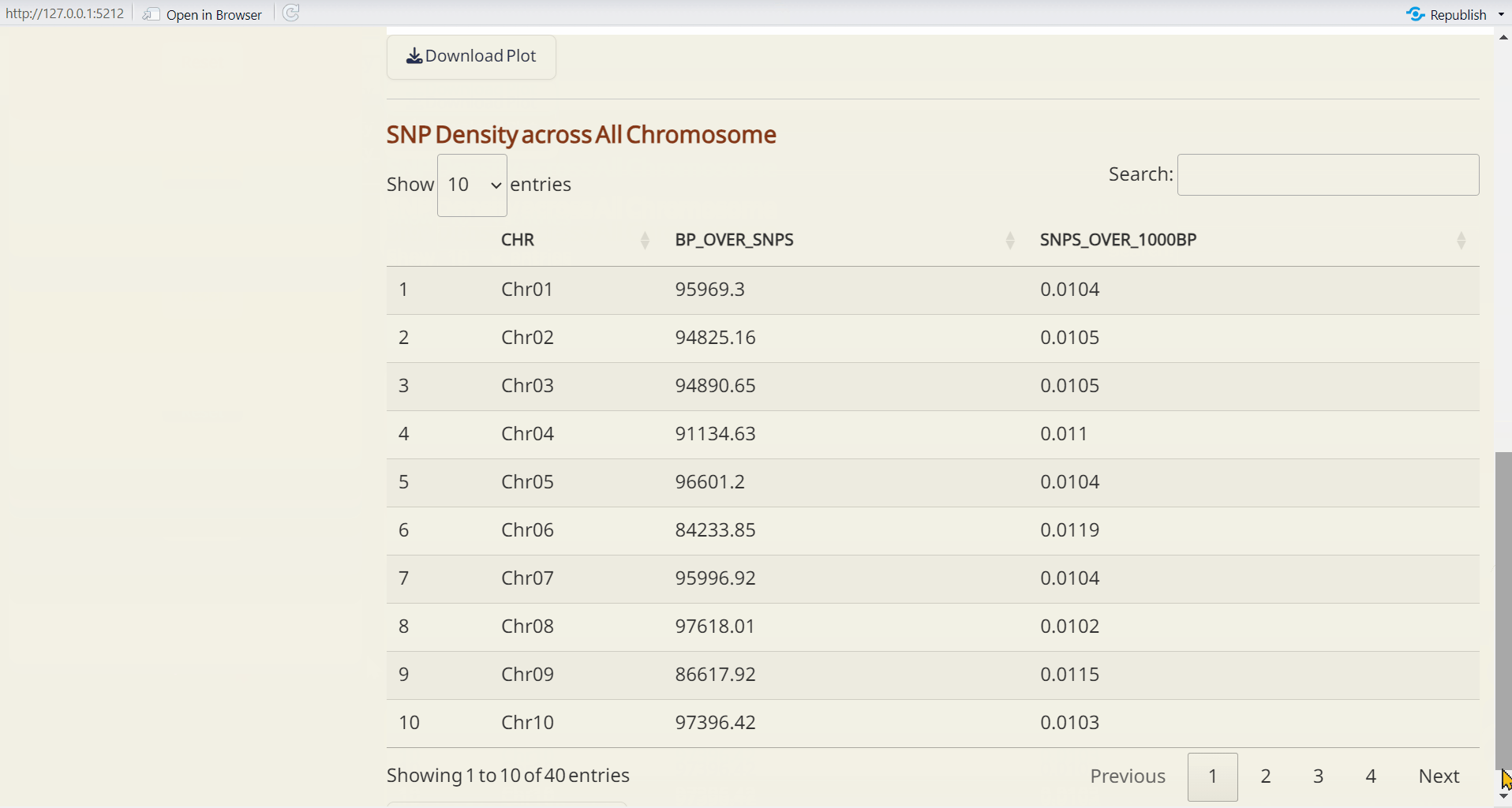
- SNP Density (CSV): A table detailing SNP density across each chromosome. bp_over_SNPs: The total base pairs (bp) per SNP in each window, representing the average spacing between SNPs. SNPs_over_1000bp: The number of SNPs per 1,000 base pairs, providing a normalized measure of SNP density across the genome.