Welcome to ShiNyP
This work, primarily authored by Yen-Hsiang Huang from the Department of Agronomy, National Chung Hsing University, Taiwan, is licensed under the GNU Affero General Public License.
Last Updated: Dec 2025
Overview
ShiNyP is a platform designed for real-time processing, analysis, and visualization of SNP datasets.
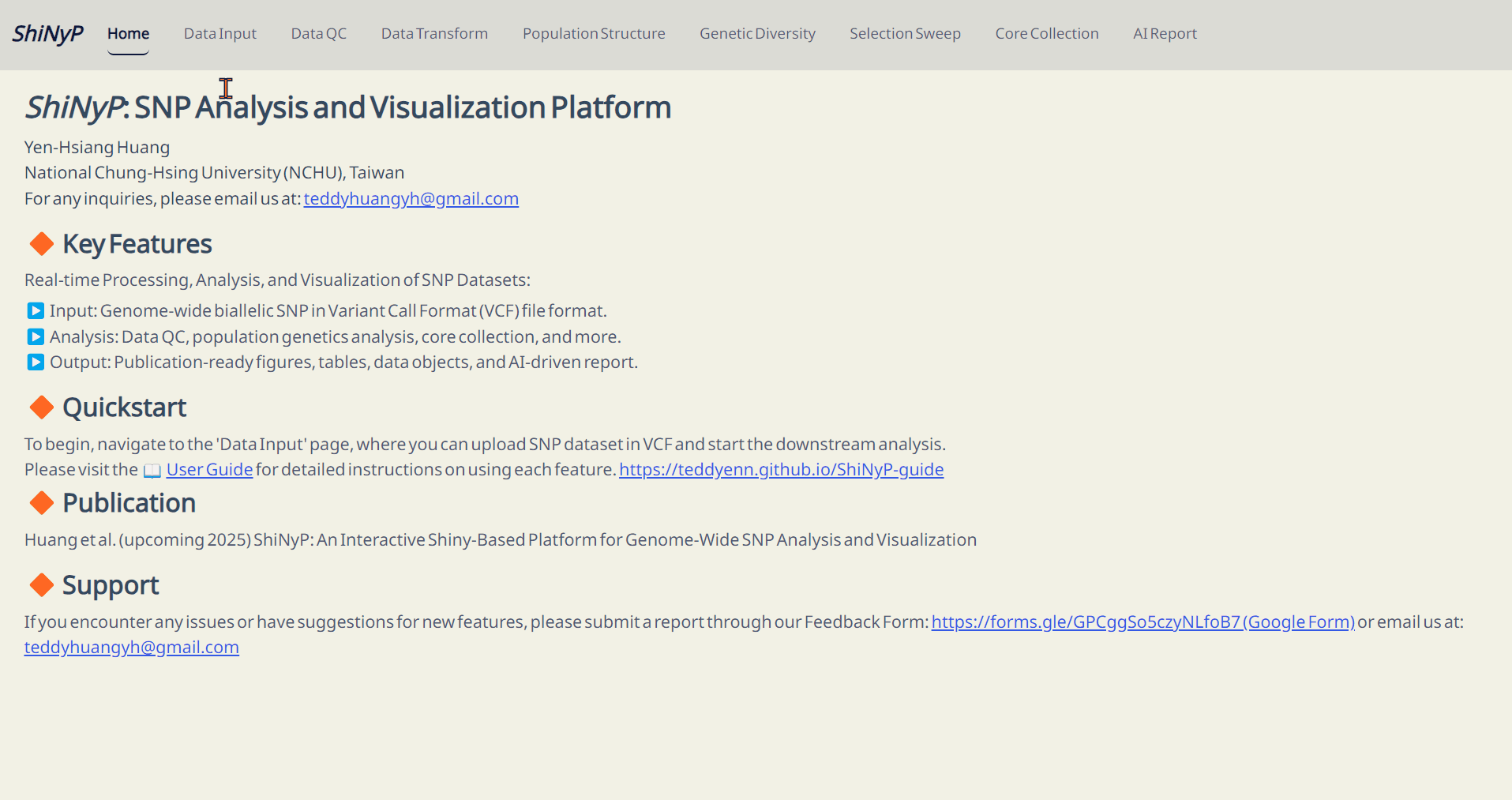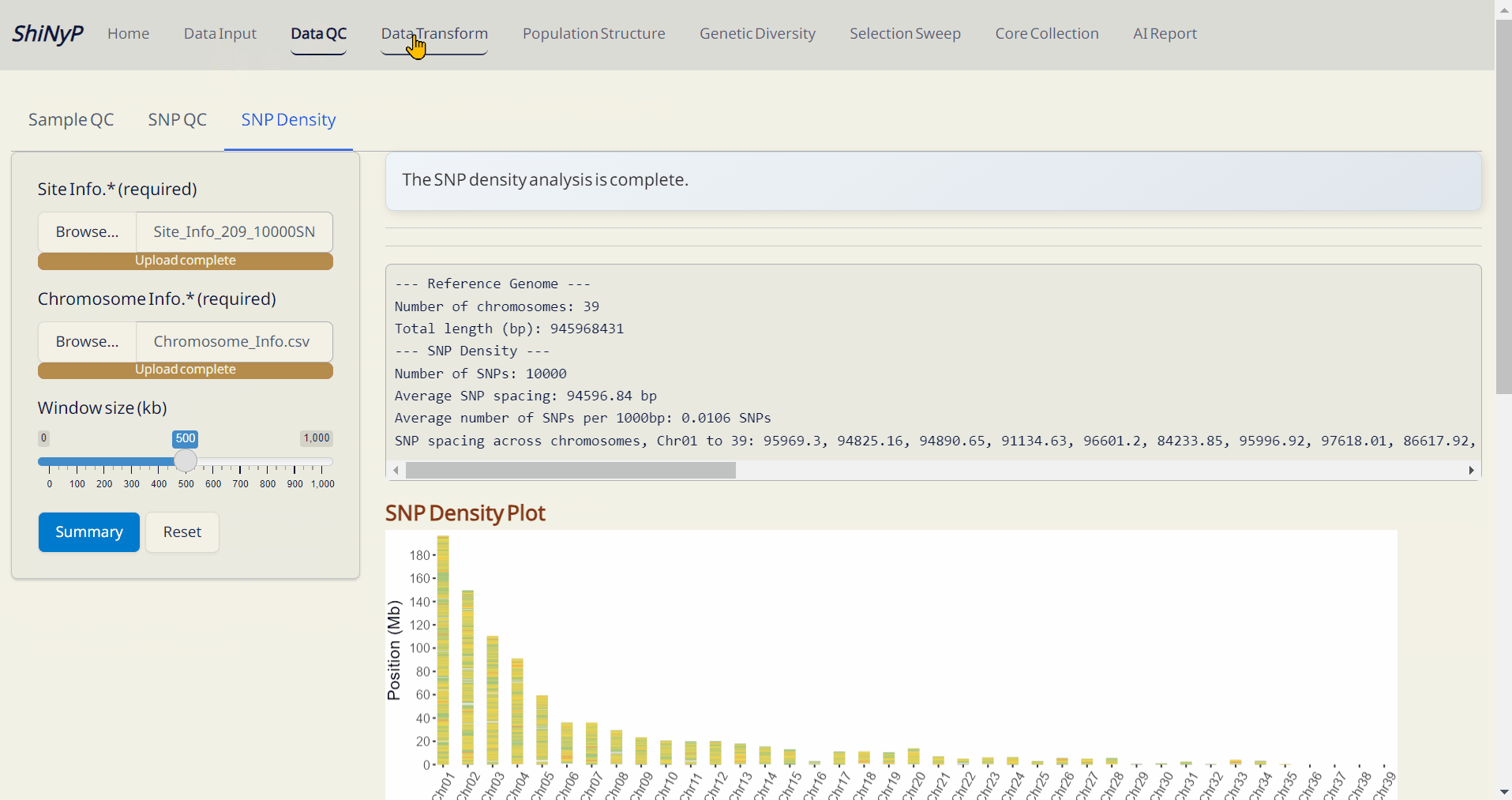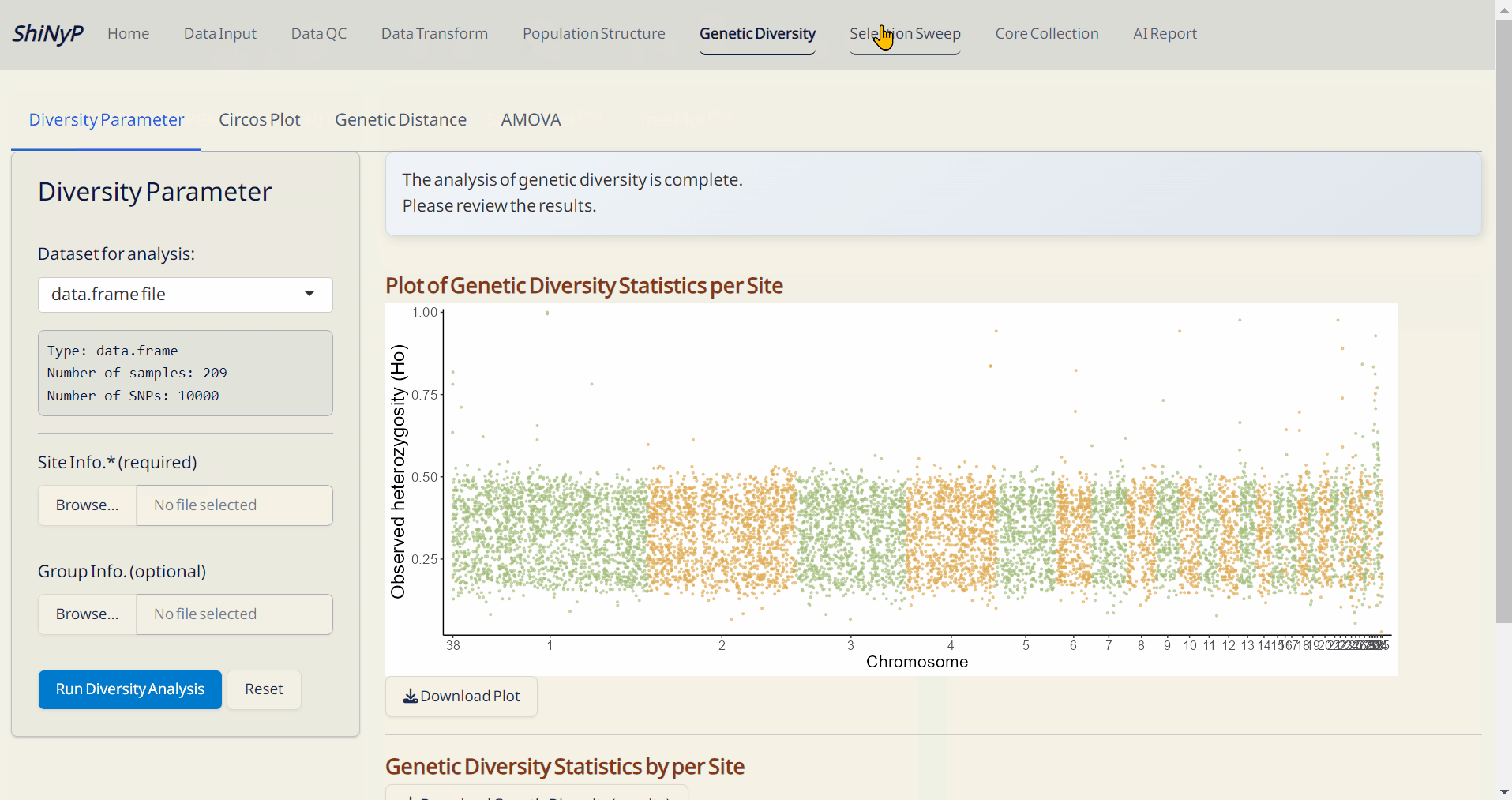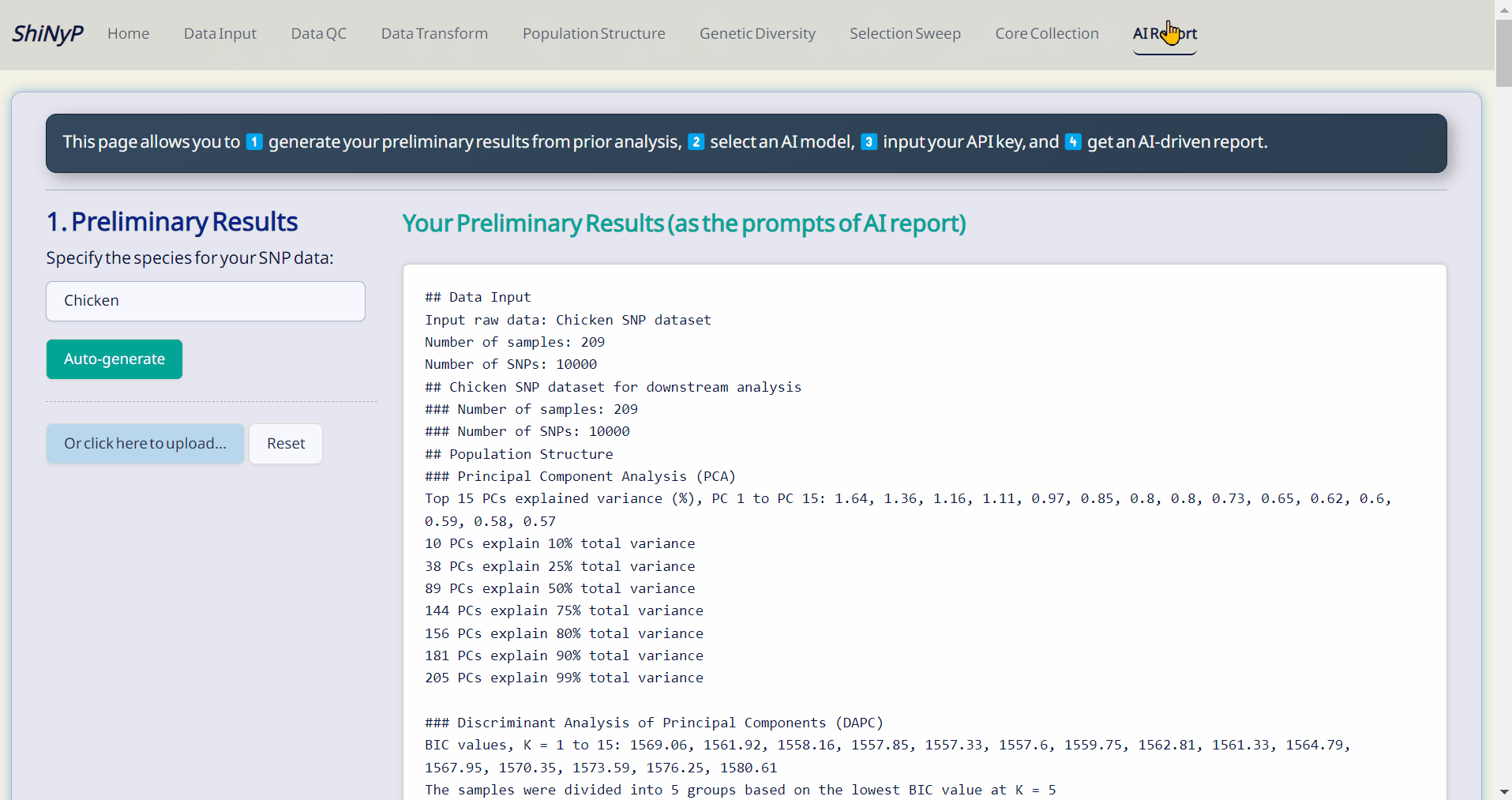
📊 Analysis: Data QC, population genetics analysis, core collection, and more.
📋 Output: Publication-ready figures, tables, R objects, and free AI-driven report.
Publication
If you use ShiNyP in your research, please cite:
Huang, Y.-H., Chen, L.-Y., Septiningsih E. M., Kao, P.-H., Kao, C.-F. (2025) ShiNyP: Unlocking SNP-Based Population Genetics—AI-Assisted Platform for Rapid and Interactive Visual Exploration. Molecular Biology and Evolution, 42(6), msaf117. https://doi.org/10.1093/molbev/msaf117
In addition, please acknowledge the R packages utilized in your analysis. The relevant citations and descriptions for each module are detailed in the ShiNyP User Guide.
Updates and Support
- Aug 2024: alpha version
- Oct 2024: v0.1.0
- Feb 2025: v0.1.1
- Apr 2025: v0.2.0
-
- Enhanced AI report functionality with new options and models.
- Improved readability of preliminary results.
- Added more methods for constructing core SNP set.
- Added the Docker-based installation. - May 2025: v1.0.0 on GitHub.
-
- Introduced the new ShiNyP AI chatbot.
- Enhanced AI report features and deprecated older AI models.
- Added publication details.
- Made UI/UX improvements.
- Fixed several bugs. - Jun 2025: v1.1.0 on GitHub.
-
- Enhanced AI report features.
- Enhanced ShiNyP AI chatbot.
- Made UI/UX improvements.
- Fixed several bugs. - 🆕 Oct 2025: v1.1.1 on GitHub.
-
- Made UI/UX improvements.
- Fixed several bugs.
- Added flexible download options.
- Added defensive & error trapping codes.
🔽 🐣 ShiNyP AI Chatbot!
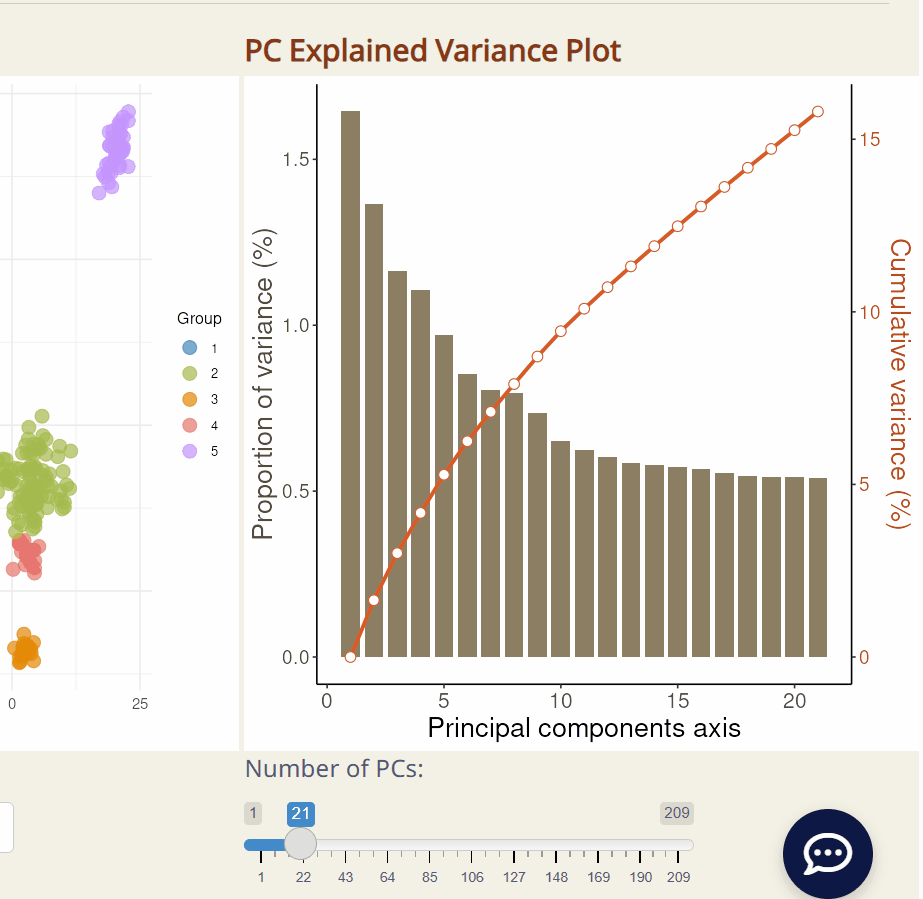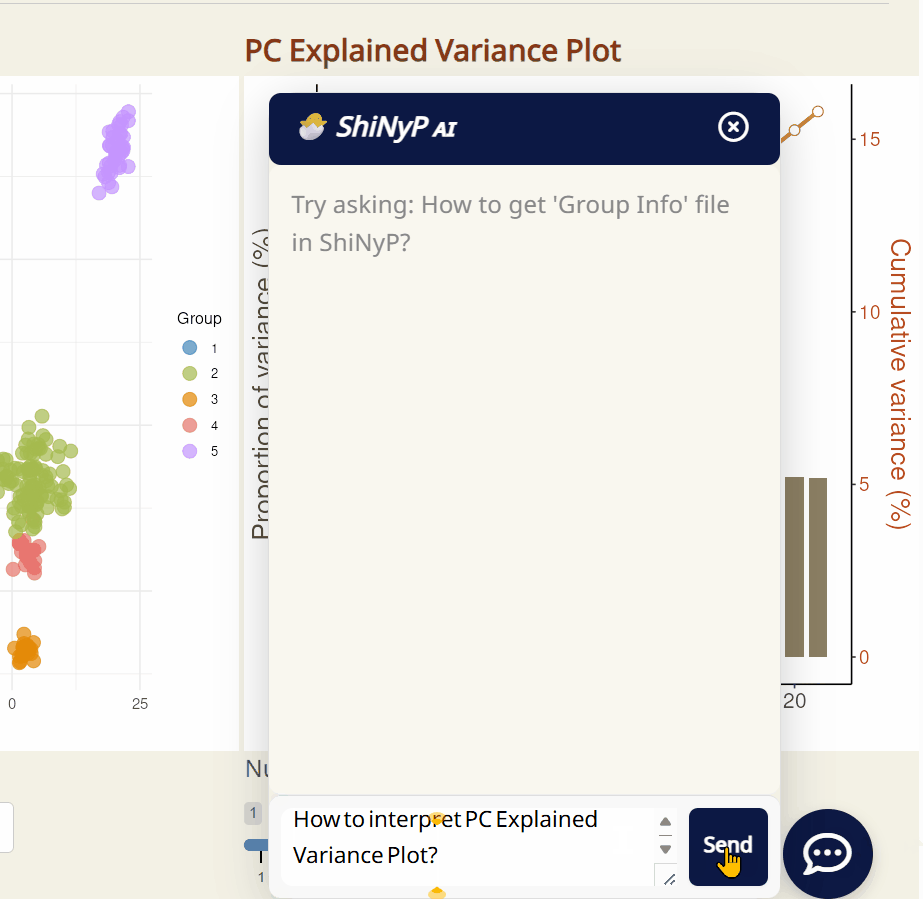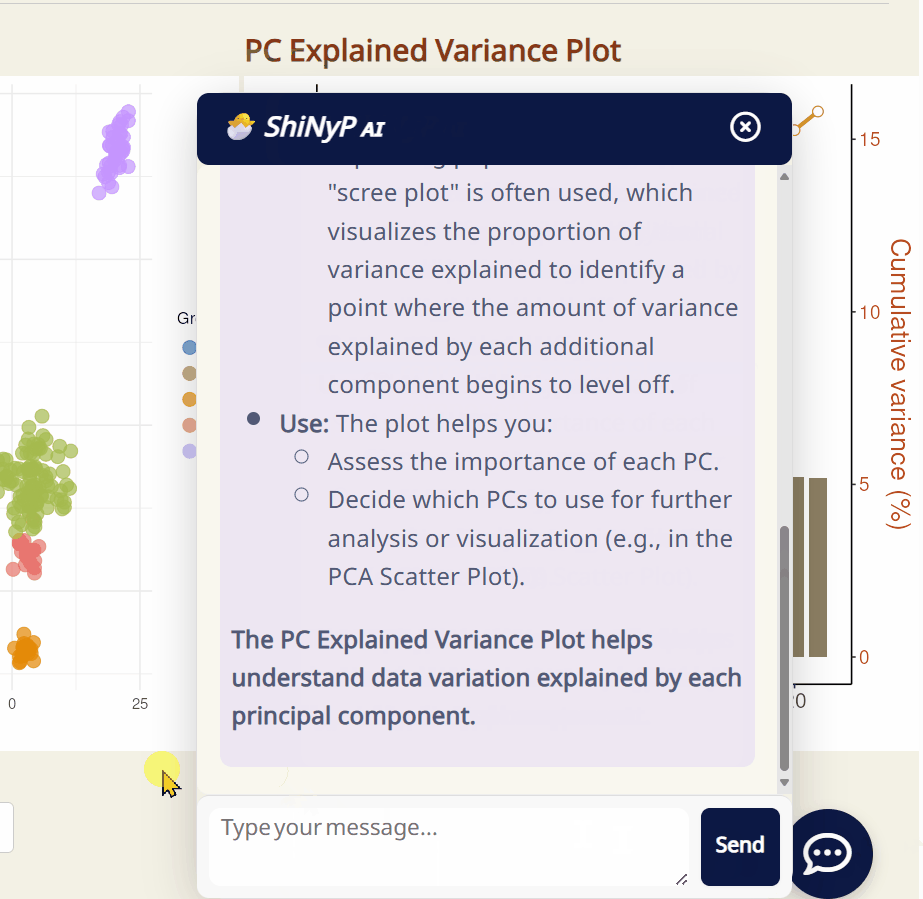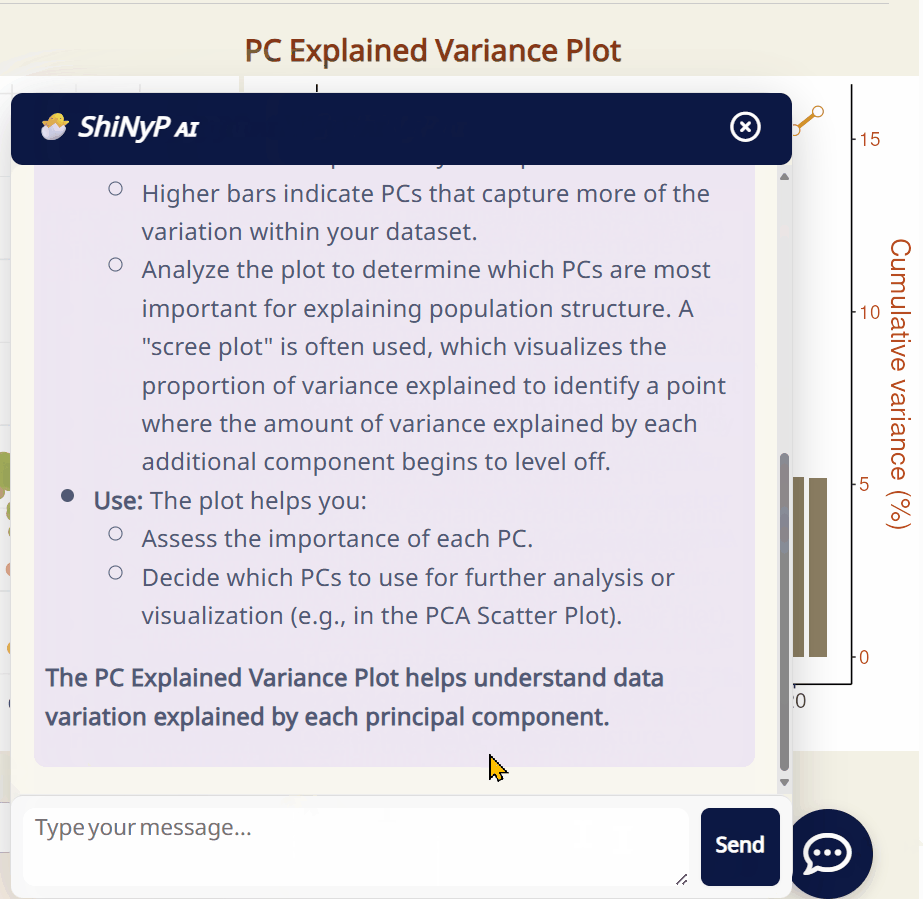
-
If you encounter any issues or have suggestions for new features, please submit a request on the GitHub Issues page or email us at: teddyhuangyh@gmail.com
This is the User Guide site for ShiNyP, live at GitHub Page.