Get Started 🚀
There are two easy ways to install and run ShiNyP:
Using R:
This method is suitable if you already have R installed or prefer working within the R environment. You’ll need to install some R packages and then launch ShiNyP directly from R environment.Using Docker:
This is an alternative if you’d rather skip installing R or any packages. With Docker, you can run ShiNyP in a ready-to-use setup with just one command.
🔘 Run ShiNyP via R
✅ Prerequisites
Before installing ShiNyP, ensure your system meets the following requirements:
-
R: Version ≥ 4.4.
Check your current version in R:
getRversion() -
Bioconductor: Version ≥ 3.20.
Match your Bioconductor version with your R version (e.g., use Bioconductor 3.21 if R = 4.5).
1️⃣ Install Required Package
install.packages("BiocManager")
BiocManager::install(version = "3.21") # Use the version that matches your R
BiocManager::install(c("qvalue", "SNPRelate", "ggtree", "snpStats"), force = TRUE)2️⃣ Install the ShiNyP Package
install.packages("remotes")
remotes::install_github("TeddYenn/ShiNyP", force = TRUE)3️⃣ Start the ShiNyP Platform
library(ShiNyP)
ShiNyP::run_ShiNyP()🔘 Run ShiNyP via Docker
If you have 🐳 Docker installed, you can launch ShiNyP without installing R.
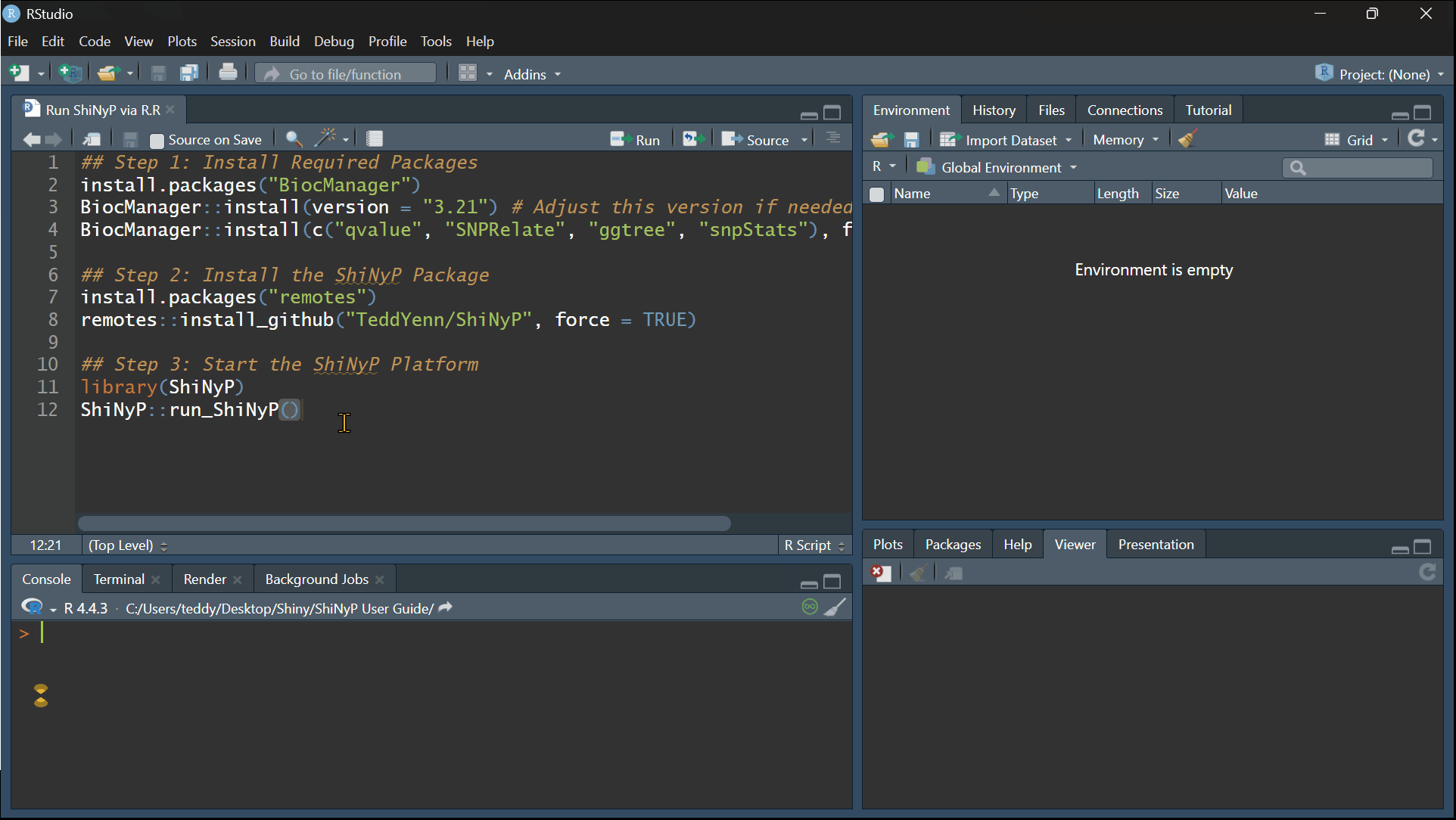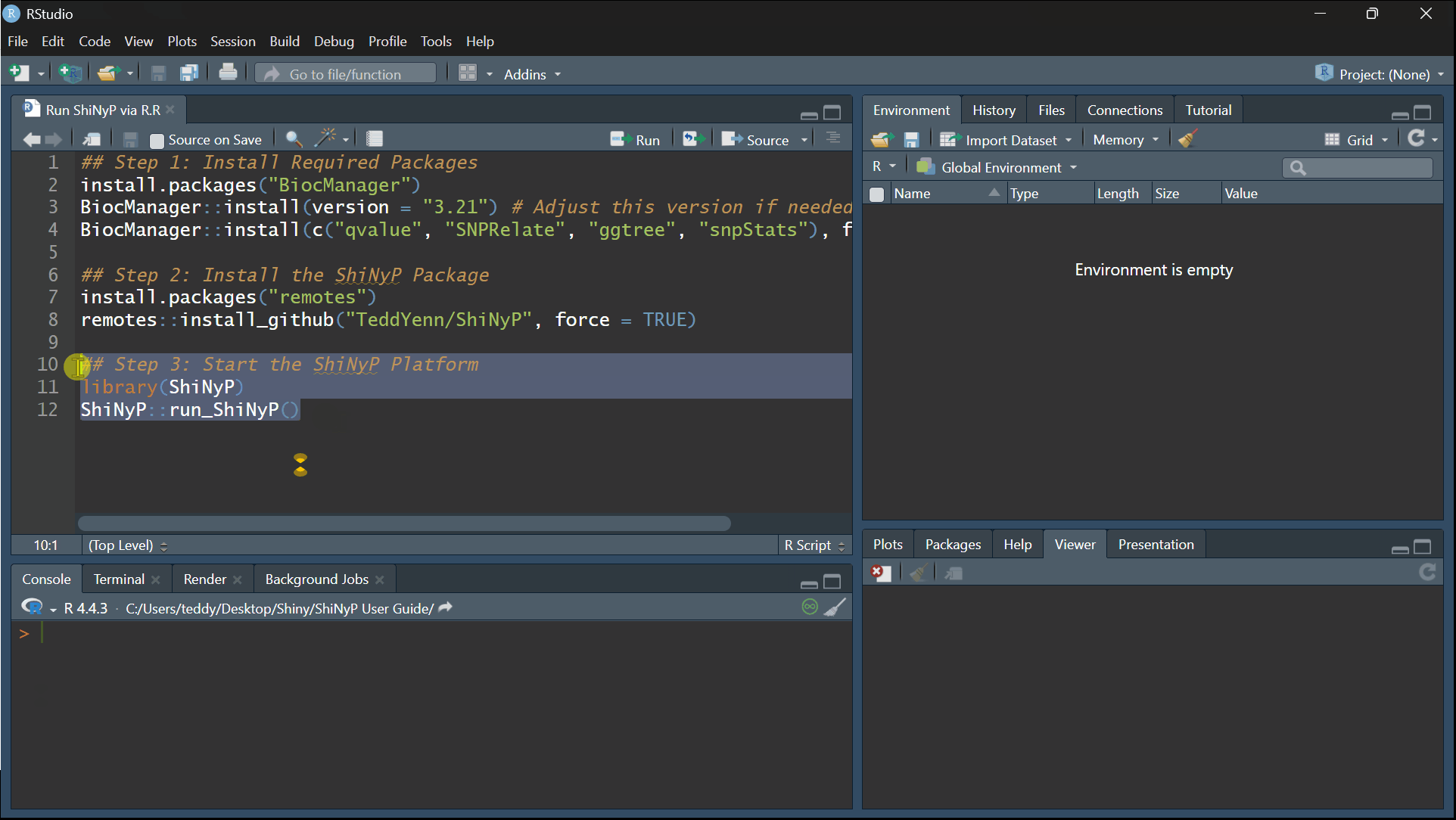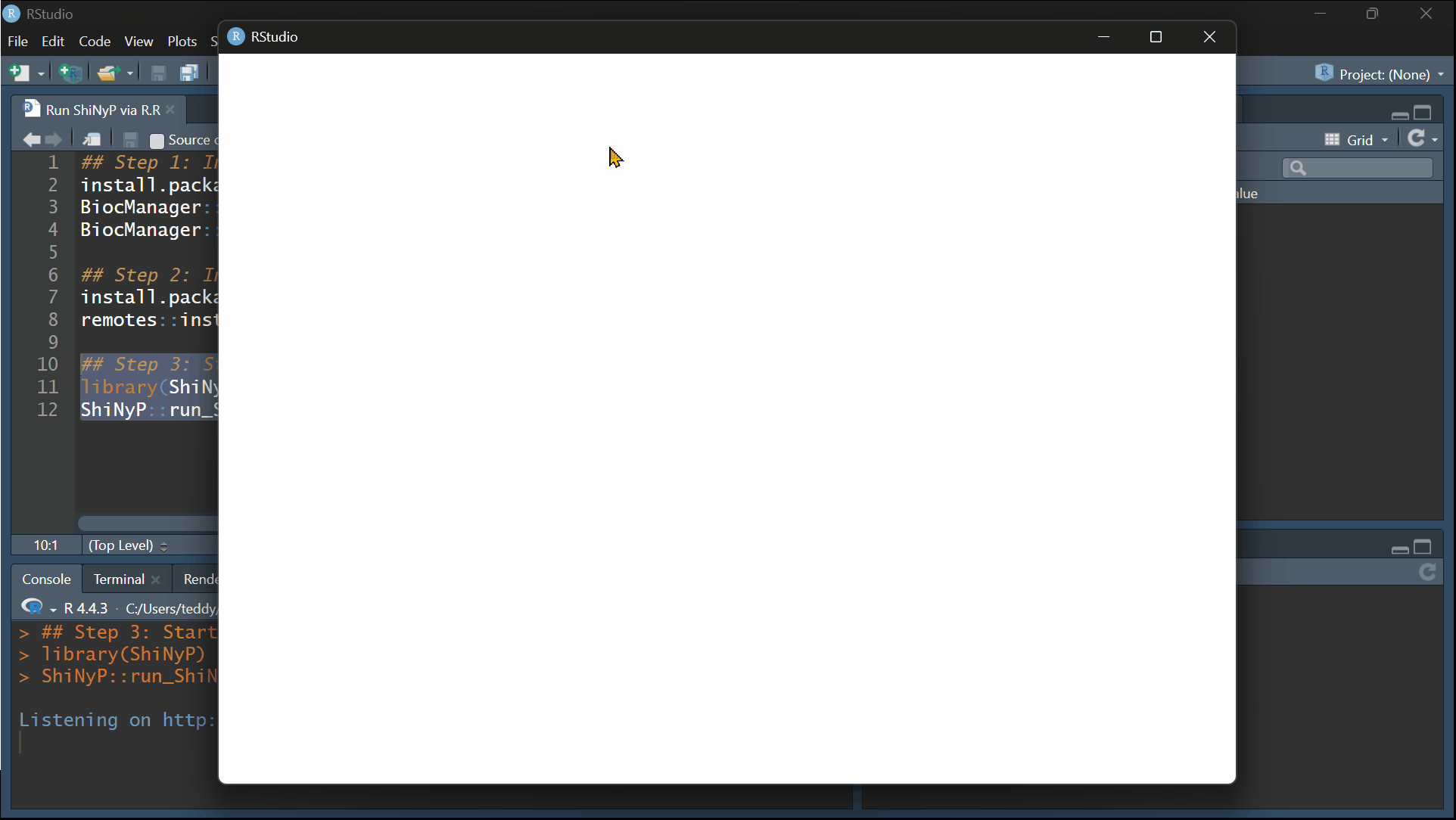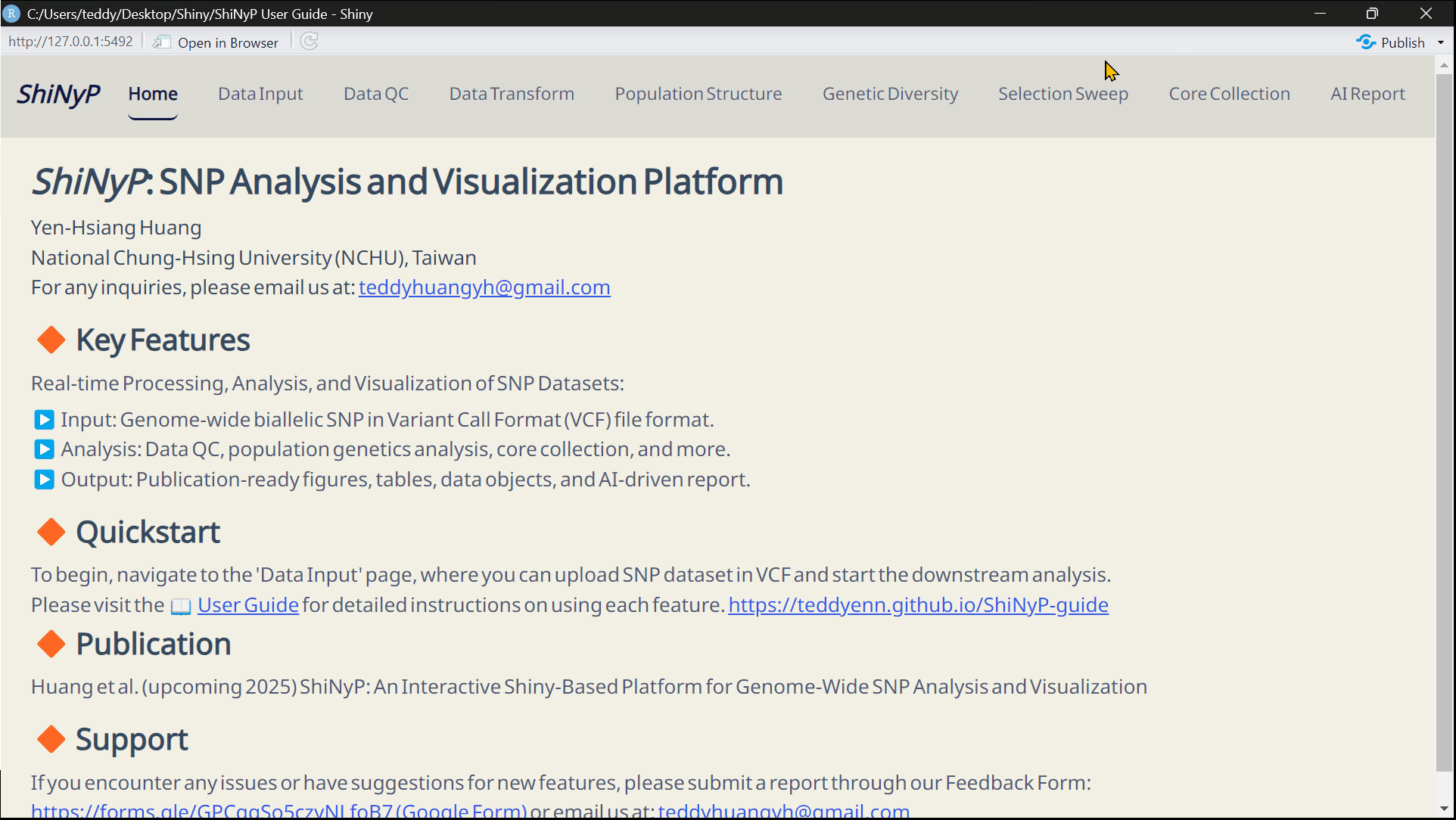
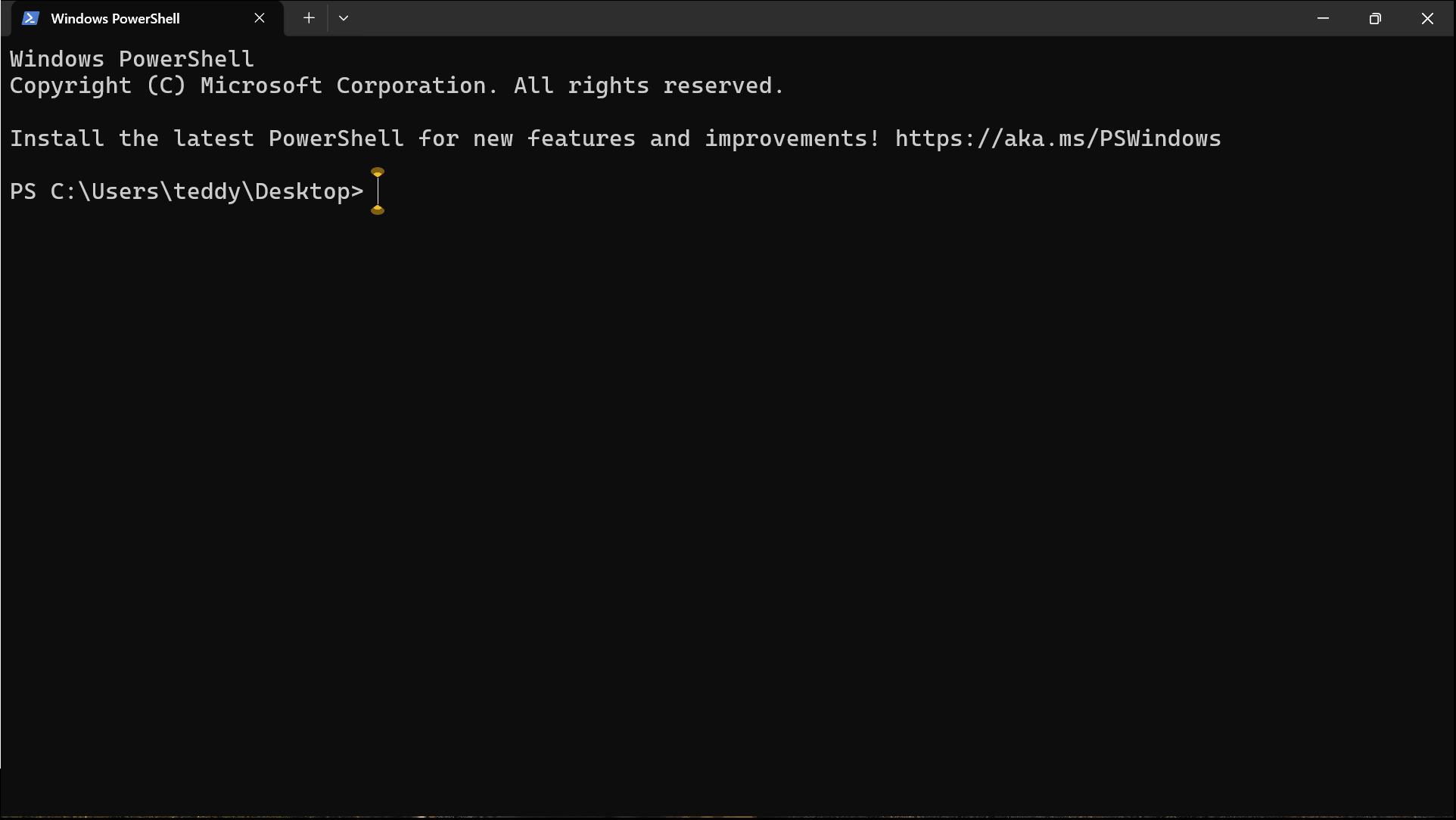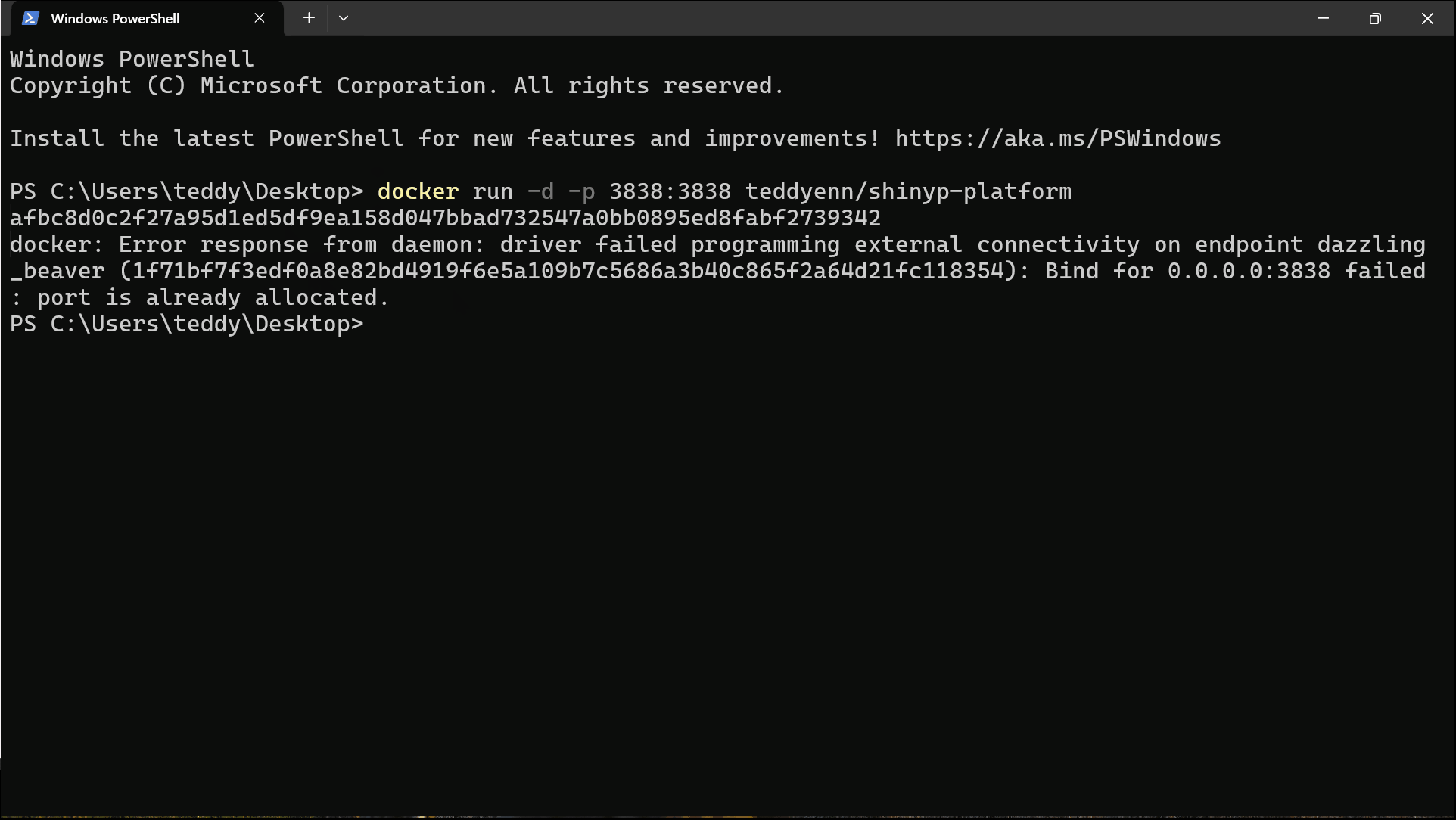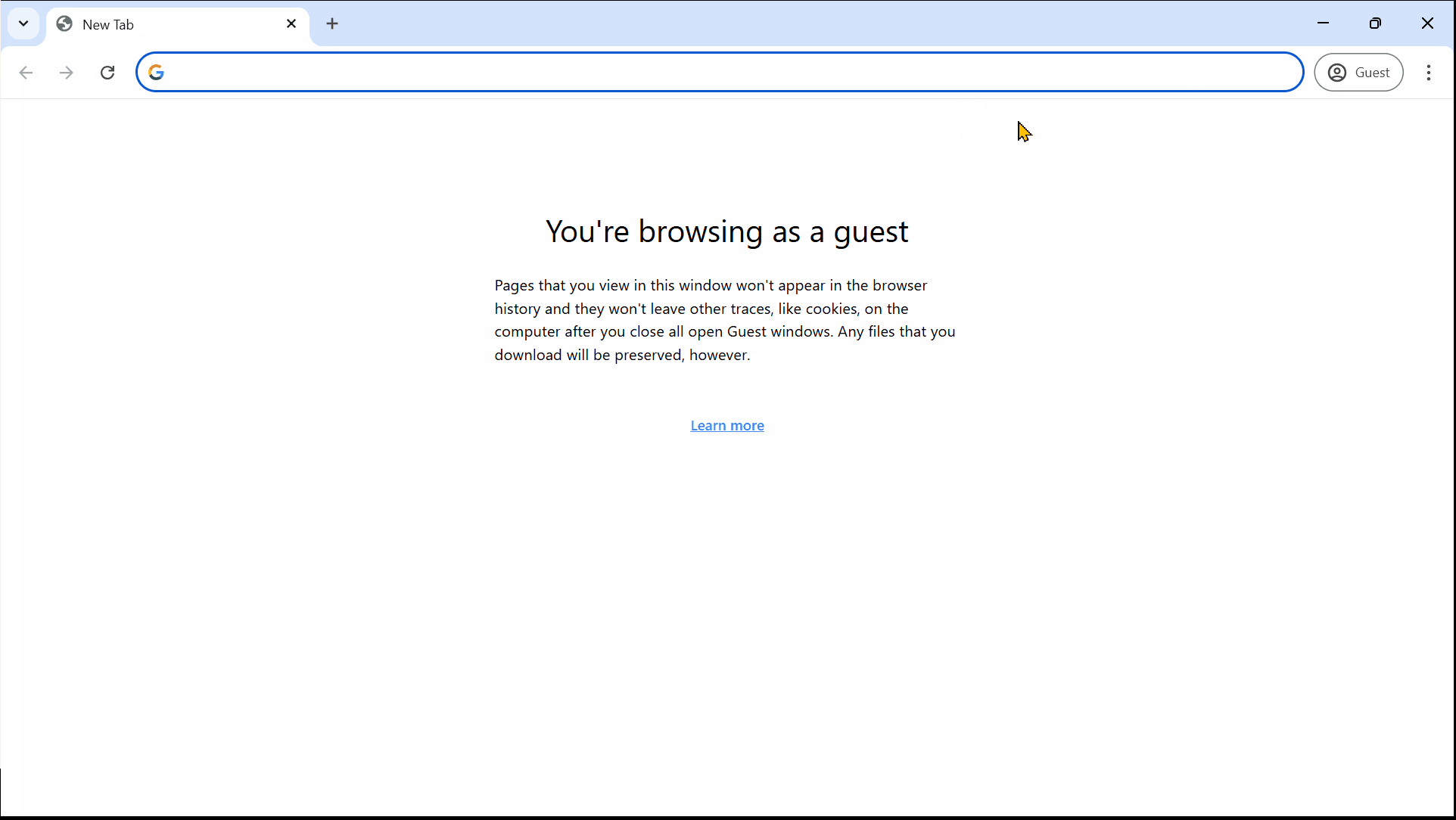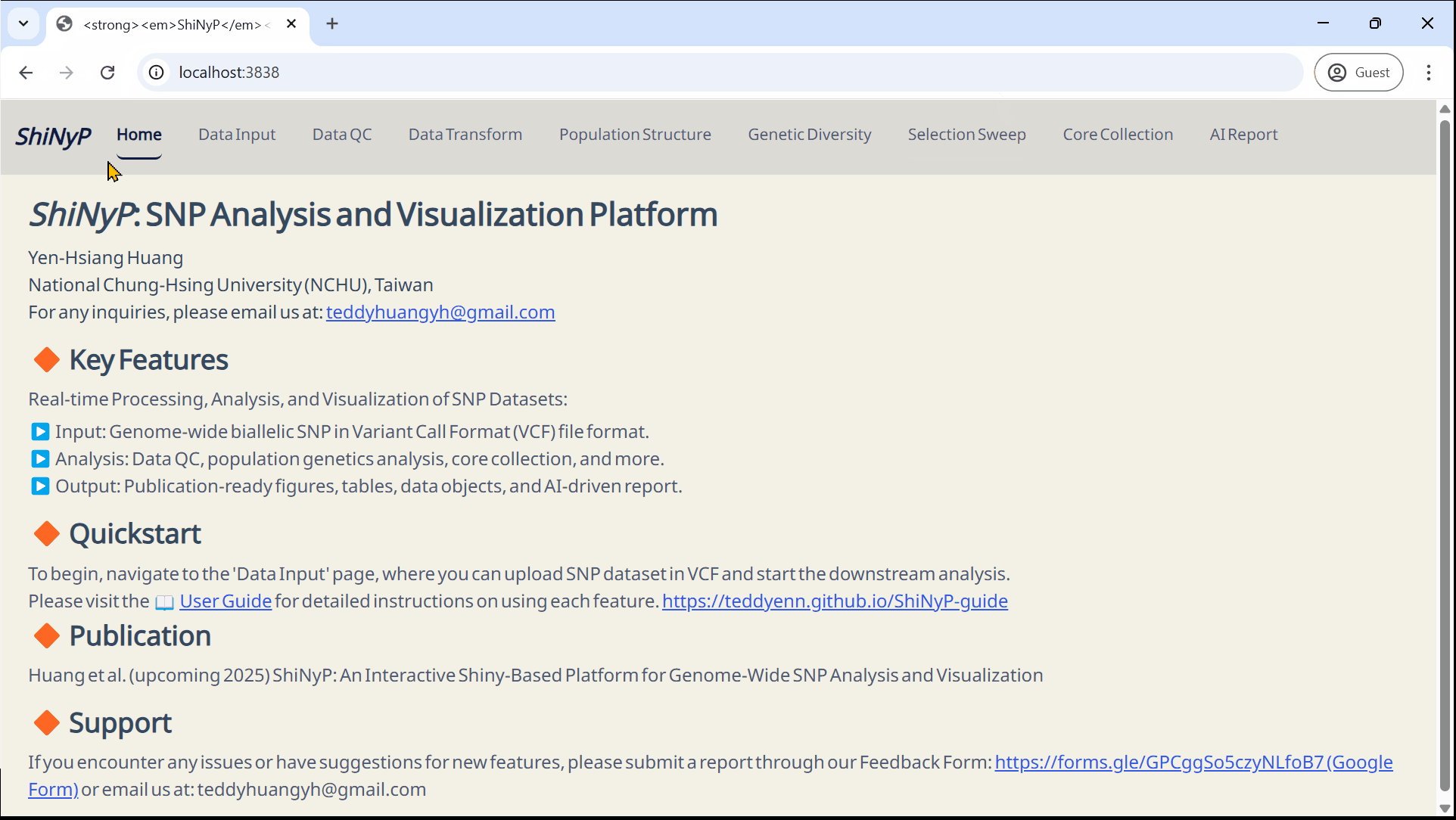
Main Features
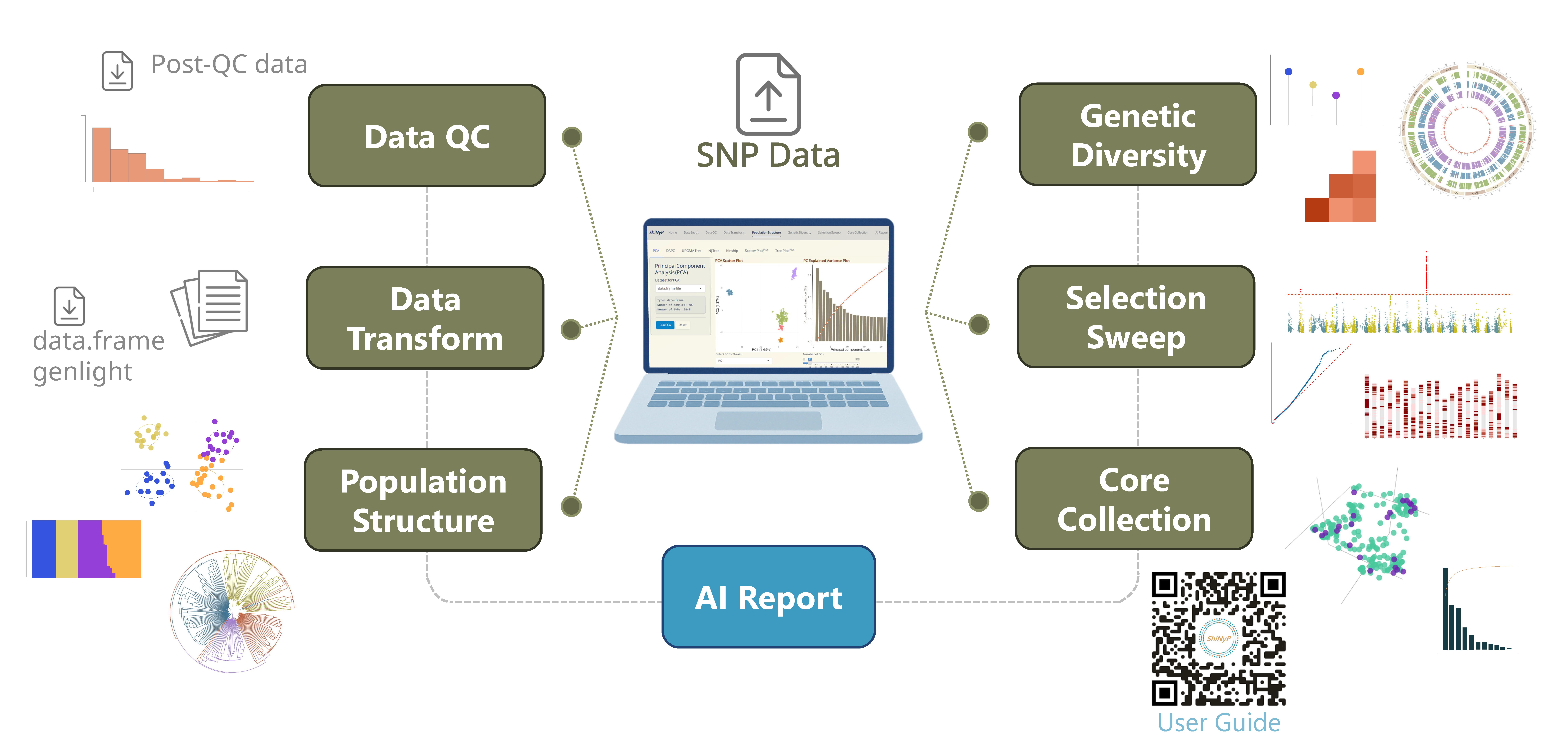
▸ Data Input & Processing:
The workflow begins with Variant Call Format (VCF) Data Input, followed by essential steps such as Data Quality Control (QC) and Data Transformation to prepare the data for analysis.
▸ Modular Analysis & Output:
Analytical functions are organized into distinct modules—each accessible as a separate page within the platform. These include: Population Structure, Genetic Diversity, Selection Sweep, and Core Collection. Each module contains multiple subpages offering specialized tools for detailed analysis.
▸ Customizable Output:
ShiNyP delivers publication-ready visualizations and AI Report that summarize analytical results in clear, structured narratives. Users can tailor output formats to fit specific research needs.