Quickstart
Prerequisites
Before installing ShiNyP, ensure your system meets the following requirements:
R Version: R ≥ 4.4 (Compatible with Bioconductor version 3.20)
R Tools (Recommended)
⚙️ Getting Started with R/RStudio/Rtools
Step 1: Install R
-
Visit the official R Project website: https://www.r-project.org/.
Windows: Click Download R for Windows, select “base,” and download the latest version.
MacOS: Click Download R for macOS and choose the correct version for your system.
Install R by double-clicking the downloaded installer and following the on-screen instructions. Use the default options unless specific needs arise.
Step 2: Install RStudio
Go to the RStudio official download page: https://posit.co/download/rstudio-desktop/.
Click Download under “RStudio Desktop - Open Source License.”
-
Select the version suitable for your operating system:
Windows:
.exefile.MacOS:
.dmgfile.
-
Locate the downloaded file and double-click it to start the installation.
Windows: Run the
.exeinstaller and follow the wizard steps.MacOS: Drag the RStudio icon into the Applications folder.
After installation, launch RStudio.
Step 3: Launch RStudio
Open RStudio by clicking the shortcut created during installation or searching for “RStudio” in your system’s application launcher.
RStudio will automatically detect your R installation and link to it.
-
RStudio has four main panels:
Console (Bottom-left): Where you run R commands.
Source (Top-left): For writing and editing scripts.
Environment/History (Top-right): Displays objects, variables, and command history.
Plots/Files/Help (Bottom-right): Displays plots, files, and R documentation.
Step 5: Install Rtools
-
Windows: Download and install the newest 64-bit Rtoolsinstaller from CRAN. Rtools is a toolchain bundle required for building R packages from source, particularly those that require C/C++ or Fortran code compilation.
Run the file to install Rtools, following the onscreen instructions. If unsure, keep the default settings. Then, return to this page and continue.
First, run
Sys.which("make"). If it detects make.exe, you’re all set. If not, configure the system path. -
MacOS: Open Terminal: Press
Cmd + Space, type “Terminal”, and pressEnter. Run the following command and pressEnter.xcode-select --installEnter your password if prompted and follow the onscreen instructions.
Verify Installation:
xcode-select -pIt should return
/Library/Developer/CommandLineTools.
Installing ShiNyP
Step 1: Pre-install Required Package
install.packages("BiocManager")
BiocManager::install(version = "3.20")
BiocManager::install(c("qvalue", "SNPRelate", "ggtree", "snpStats"), force = TRUE)Step 2: Install the ShiNyP Package from GitHub
install.packages("remotes")
remotes::install_github("TeddYenn/ShiNyP", force = TRUE)Step 3: Start the ShiNyP Platform
library(ShiNyP)
ShiNyP::run_ShiNyP()Step 4: Run ShiNyP Analysis
Input your SNP data in VCF format, or feel free to use our Demo Data.
Note: If you run in RStudio, you can click the Open in Browser button.
For any inquiries, please email us at: teddyhuangyh@gmail.com.
Troubleshooting
During installation or while running ShiNyP, you might encounter issues. Below are common problems and their solutions.
- ⚒️ R and Bioconductor Version Mismatch
-
The installation specifies Bioconductor version 3.19, which requires R version ≥ 4.4. If your R version is below 4.4, update it from CRAN.
# Check your R version R.version.string
- ⚒️ Installation of Dependencies
-
Installing packages like shiny and dartR may require additional developer tools. You might encounter error messages such as:
-
-
For Windows:
Download Rtools from CRAN Rtools.
Run the installer and follow the on-screen instructions.
Ensure Rtools is added to your system PATH during installation.
Verify Installation:
pkgbuild::find_rtools()It should return
TRUEif Rtools is correctly installed. -
For macOS:
Open Terminal: Press
Cmd + Space, type “Terminal”, and pressEnter.Run the Installation Command:
xcode-select --installVerify Installation:
xcode-select -pIt should return
/Library/Developer/CommandLineTools. -
For Linux:
Install Essential Build Tools:
-
- ⚒️ GitHub Installation Issues
-
Verify that your system is connected to the internet. Check for any firewall or proxy settings that might block GitHub access.
- ⚒️ Package Loading Issues
-
Errors occur when loading ShiNyP or its dependencies, possibly due to outdated packages or conflicts.
-
Update All Installed Packages:
When prompted with
# These packages have more recent versions available. # It is recommended to update all of them. # Which would you like to update? # -> Respond with 1 to update all packages.Please type 1 (1: All)
-
If a specific package is causing issues, reinstall it.
install.packages("dartRverse") BiocManager::install("SNPRelate") -
Check for Namespace Conflicts:
Identify if multiple packages are masking functions.
-
Main Functions
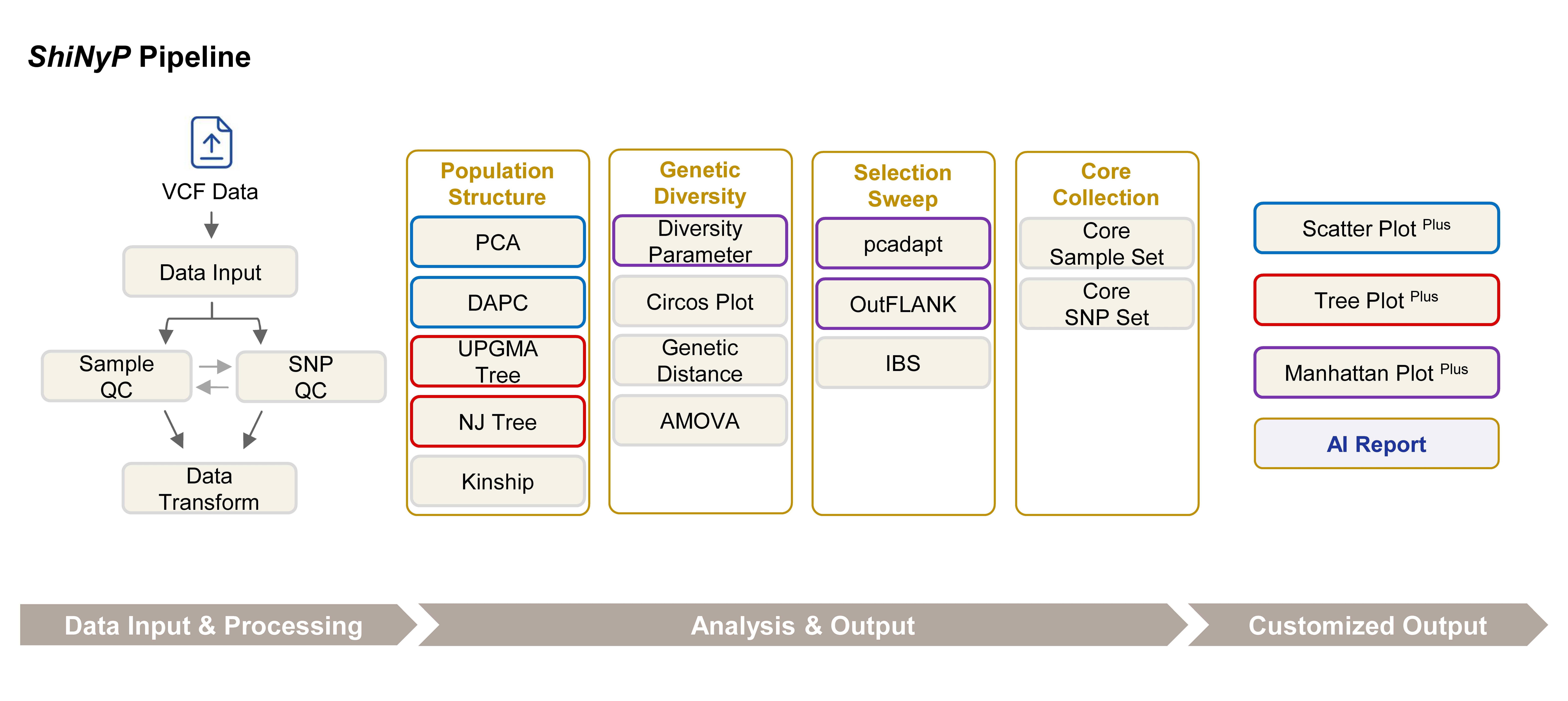
▶️ Data Input & Processing: Beginning with VCF data input, it performs quality control (QC) and data transformation steps.
▶️ Analysis & Output: The analysis phase is divided into modules, each represented as a page in the platform, with multiple subpages offering specific analytical functions, including population structure, genetic diversity, selection sweep, and core collection.
▶️ Customized Output: The final output offers tailored visualizations and includes an AI-generated report summarizing the results. The pipeline streamlines data input, processing, and advanced analysis to deliver publication-ready figures and reports customized to the user’s needs.
*Subpage frame colors indicate available functions for customization. For example, blue frames for PCA and DAPC correspond to the Scatter Plot Plus tool for customizing scatter plots, while red and purple frames correspond to Tree Plot Plus and Manhattan Plot Plus, respectively.