1 Introduction
Soybeans (Glycine max L.)
are dichotomously categorized into grain and vegetable utilization types based on their developmental stage at the time of harvest. Grain soybeans are conventionally harvested at reproductive stage 8 (R8). Vegetable soybeans, commonly known as edamame, are typically harvested at stage R6. Vegetable soybeans boast large seed sizes and are rich sources of plant proteins, carbohydrates, dietary fiber, vitamins, minerals, antioxidants, and isoflavones (Kumar et al. 2011). The rising awareness of healthy dietary pattern has driven the demand for vegetable soybeans across diverse geographic regions, including Asia, the Americas, Europe, and sub-Saharan Africa (Huang et al. 2022; Nair et al. 2023). Soybeans were domesticated from wild soybeans (Glycine soja Sieb. & Zucc.) is estimated to have occurred as early as 6,000-9,000 years ago in China (Sedivy et al. 2017), and soybeans were already being consumed as a vegetable around 1,000-2,200 years ago (Shanmugasundaram et al. 1991). Contemporary statistics depict a global surge in vegetable soybean cultivation, with China, Japan, and Taiwan emerging as pivotal centers of production (Nair et al. 2023).
Most soybean research
focused on grain soybeans due to the extensive cultivation to serve a wide range of purposes, less attention has been given to vegetable soybeans (Nair et al. 2023). The diverse breeding objectives and selection criteria for vegetable and grain soybeans likely led to the development of distinct genetic profiles (Liu et al. 2022; Viana et al. 2022). These unique selection pressures have played a significant role in shaping the genetic diversity and differentiation of vegetable and grain soybeans throughout variety improvement programs (Lu et al. 2020; Mendonça et al. 2022). To devise more precise and efficient breeding strategies, it is crucial to have a comprehensive understanding of the genetic similarities or distinctions regulating desirable characteristics between two types of soybeans.
In recent years,
research efforts in the field of vegetable soybeans have primarily focused on identifying genetic loci linked to key agronomic traits such as seed weight, pod weight, and soluble sugar content (Li et al. 2019; Lu et al. 2022). While these investigations have significantly advanced our understanding of trait-specific genetic markers in vegetable soybeans, there remains a notable gap in our exploration of the genetic background, with few studies directed toward investigating the population structure of vegetable soybeans (Nair et al. 2023). Alternatively, some studies have only broadly categorized the germplasms into soybeans, vegetable soybeans, and wild soybeans for research, potentially oversimplifying the genetic makeup (Liu et al. 2022). To bridge this gap, future research should employ advanced genomic tools to analyze a larger number of genetic markers across diverse populations. Additionally, the sampling strategy of germplasms should be expanded to encompass regions with distinct environmental conditions to capture the full spectrum of genetic diversity. The incorporation of the rich reservoirs of germplasm from Taiwan and Japan (Huang et al. 2022; Nair et al. 2023) would provide valuable insights into the population structure and genetic architecture of vegetable soybeans.
The National Plant Genetic Resources Center (NPGRC)
at the Taiwan Agricultural Research Institute houses a broad collection of plant germplasms, comprising a diverse panel of soybean accessions sourced from domestic and exotic landraces and varieties. Within this collection, certain ancestors vegetable soybeans in Taiwan have been incorporated into breeding programs (Huang et al. 2022). A notable example is the ‘Ryokkoh’ from Japan, a superior ancestor of vegetable soybean that has been used as a parent genotype for several decades. This lineage has contributed to the development of numerous elite hybrid lines, including Kaohsiung No. 2, 5, 8, 9, and 13 (Chou 2015). Consequently, the germplasm in Taiwan not only provides extensive and diverse resources but also preserves promising genotypes possessing desired traits. To facilitate a more accurate comparison of the genetic differences between grain and vegetable soybeans and to identify selection footprints specifically in vegetable soybeans, we have established a core collection comprising both types of soybeans. This approach could help to mitigate potential biases that may arise from differences in sample sizes and the representation of germplasm. By establishing a core collection, we can more fairly evaluate the genetic diversity, population structure, and trait variability between these two types of soybeans.
The current study
introduced a novel pipeline aimed at identifying selection footprints by utilizing a core collection framework of Taiwanese vegetable and grain soybean germplasms (Fig. 1). Integrating large-scale genotypic and phenotypic data, we elucidated the genomic architecture, genetic similarities or differences, and variations of core accessions between two soybean types. Additionally, a genome-wide scan was conducted to identify distinct genetic signatures and selection signatures, providing insight into the genetic basis of favorable or novel alleles in vegetable soybeans based on the core collection framework. This approach is pivotal for guiding breeding programs and developing cultivars with improved agronomic traits tailored to local environments and market demands.
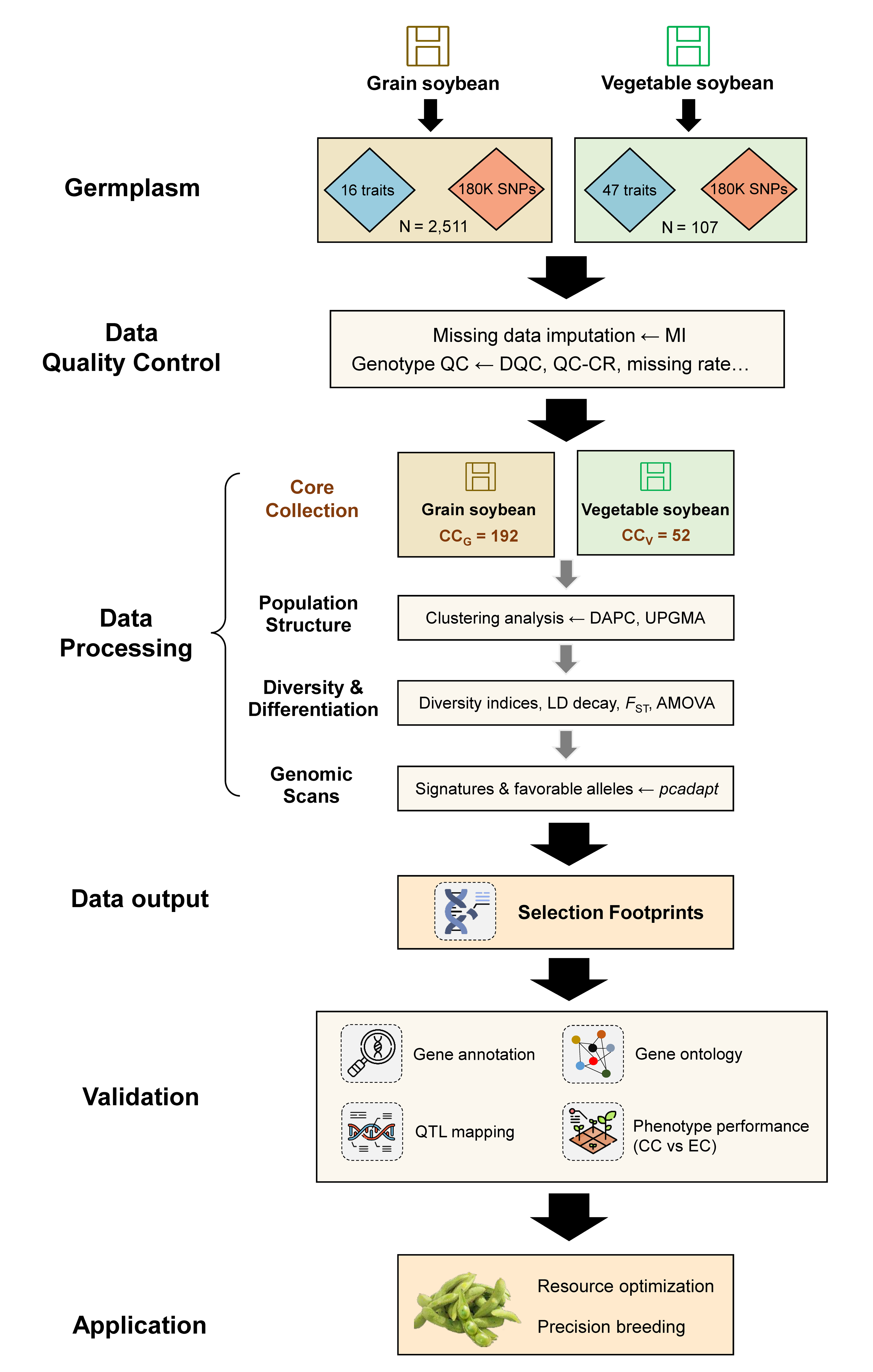
Fig. 1 A novel pipeline using core collection framework for selection footprints in vegetable soybeans. The soybean germplasm comprising 2,618 Taiwanese soybean accessions, underwent phenotypic evaluation and Axiom® SoyaSNP180K chip array genotyping. Data quality control was implemented to address missing phenotypic data through multiple imputation (MI), while poor-quality samples and markers were filtered out in single nucleotide polymorphism (SNP) datasets. Core collections were established for both grain (CCG) and vegetable (CCV) soybeans, from the entire collections of grain (ECG) and vegetable (ECV), respectively, with phenotypic and genotypic datasets. Data analyses were conducted to evaluate population structure, genetic diversity and differentiation. Furthermore, genome-wide scans were performed to pinpoint loci indicative of selection footprints in vegetable soybeans. The validation step involved examining gene function, QTL mapping (GWAS QTLs data), and phenotypic trait to validate the identified loci, genes, and genomic regions. Prospective applications of this methodology hold promise for advancing vegetable soybean breeding and enhancement strategies through targeted selection. DAPC, discriminant analysis of principal components; UPGMA, unweighted pair group method with arithmetic mean; LD, linkage disequilibrium; AMOVA, analysis of molecular variance; QTL, quantitative trait locus